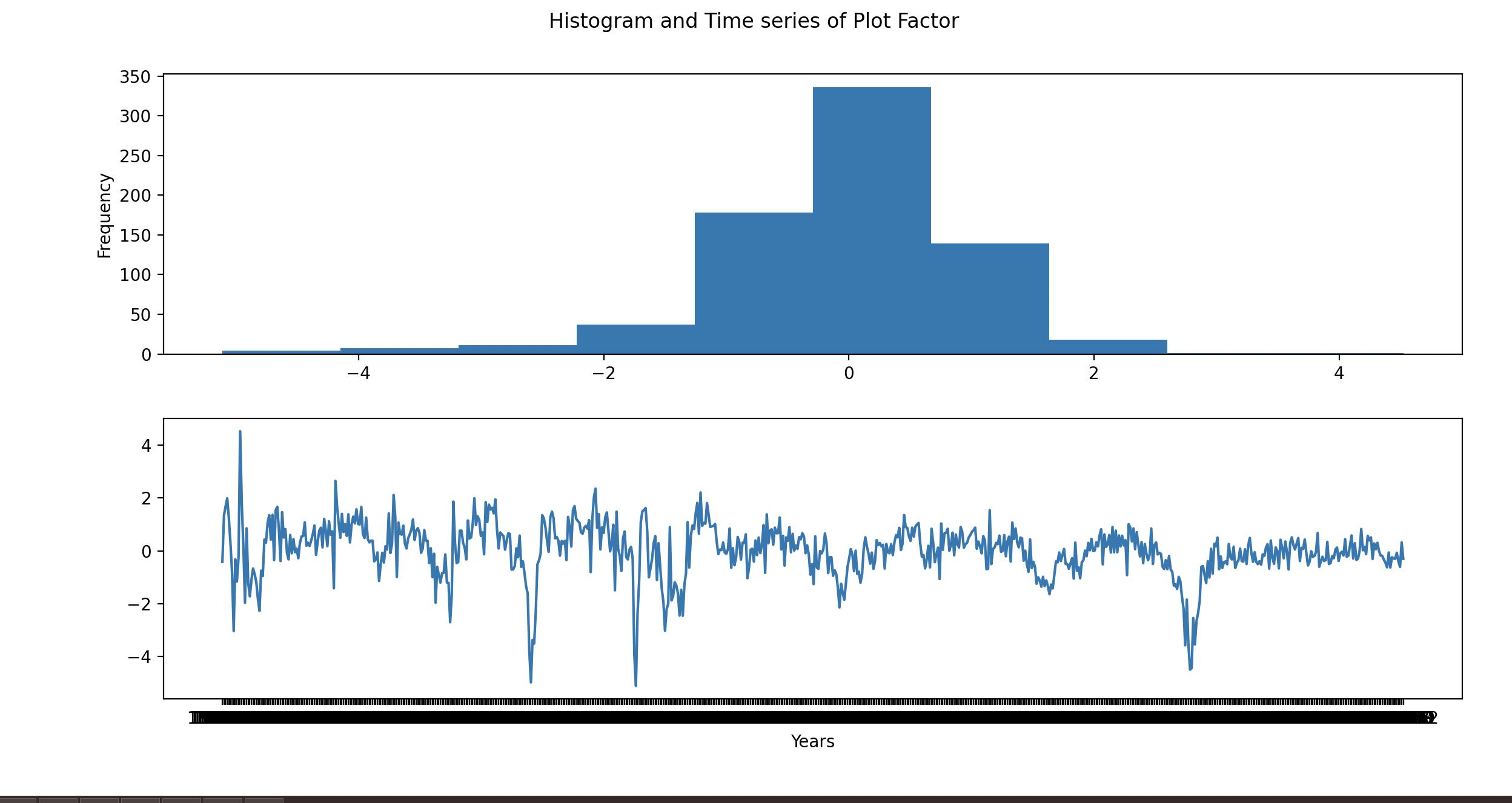
I'm trying to plot a graph of a time series which has dates from 1959 to 2019 including months, and I when I try plotting this time series I'm getting a clustered x-axis where the dates are not showing properly. How is it possible to remove the months and get only the years on the x-axis so it wont be as clustered and it would show the years properly?
fig,ax = plt.subplots(2,1)
ax[0].hist(pca_function(sd_Data))
ax[0].set_ylabel ('Frequency')
ax[1].plot(pca_function(sd_Data))
ax[1].set_xlabel ('Years')
fig.suptitle('Histogram and Time series of Plot Factor')
plt.tight_layout()
# fig.savefig('factor1959.pdf')
pca_function(sd_Data)
comp_0
sasdate
1959-01 -0.418150
1959-02 1.341654
1959-03 1.684372
1959-04 1.981473
1959-05 1.242232
...
2019-08 -0.075270
2019-09 -0.402110
2019-10 -0.609002
2019-11 0.320586
2019-12 -0.303515
[732 rows x 1 columns]
CodePudding user response:
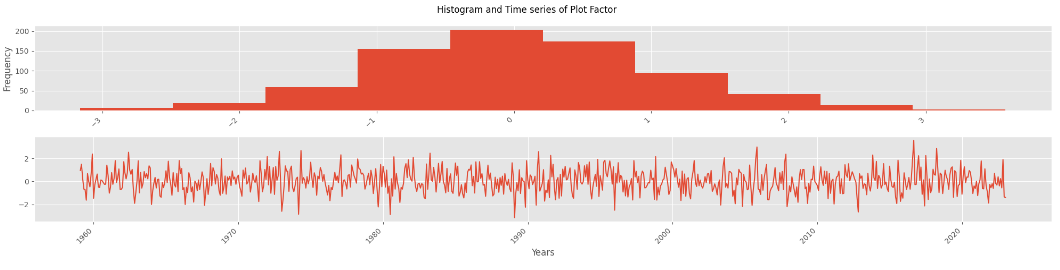
From what I see, you do have years on your second subplot, they are just overlapped because there are to many of them placed horizontally. Try to increase figsize, and rotate ticks:
# Builds an example dataframe.
df = pd.DataFrame(columns=['Years', 'Frequency'])
df['Years'] = pd.date_range(start='1/1/1959', end='1/1/2023', freq='M')
df['Frequency'] = np.random.normal(0, 1, size=(df.shape[0]))
fig, ax = plt.subplots(2,1, figsize=(20, 5))
ax[0].hist(df.Frequency)
ax[0].set_ylabel ('Frequency')
ax[1].plot(df.Years, df.Frequency)
ax[1].set_xlabel('Years')
for tick in ax[0].get_xticklabels():
tick.set_rotation(45)
tick.set_ha('right')
for tick in ax[1].get_xticklabels():
tick.set_rotation(45)
tick.set_ha('right')
fig.suptitle('Histogram and Time series of Plot Factor')
plt.tight_layout()
p.s. if the x-labels still overlap, try to increase your step size.
CodePudding user response:
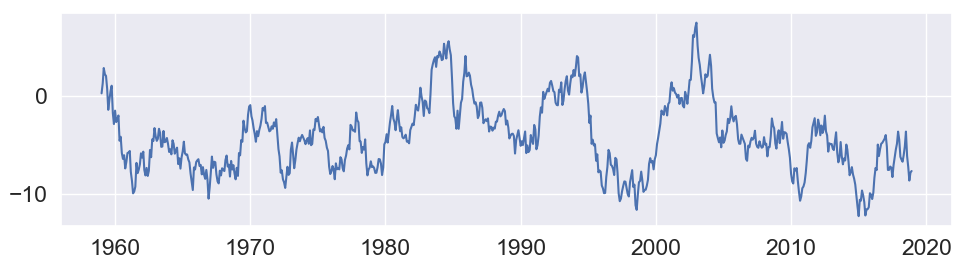
First off, you need to store the result of the call to pca_function into a variable. E.g. called result_pca_func. That way, the calculations (and possibly side effects or different randomization) are only done once.
Second, the dates should be converted to a datetime format. For example using pd.to_datetime(). That way, matplotlib can automatically put year ticks as appropriate.
Here is an example, starting from a dummy test dataframe:
import matplotlib.pyplot as plt
import pandas as pd
import numpy as np
df = pd.DataFrame({'Date': [f'{y}-{m:02d}' for y in range(1959, 2019) for m in range(1, 13)]})
df['Values'] = np.random.randn(len(df)).cumsum()
df = df.set_index('Date')
result_pca_func = df
result_pca_func.index = pd.to_datetime(result_pca_func.index)
fig, ax2 = plt.subplots(figsize=(10, 3))
ax2.plot(result_pca_func)
plt.tight_layout()
plt.show()