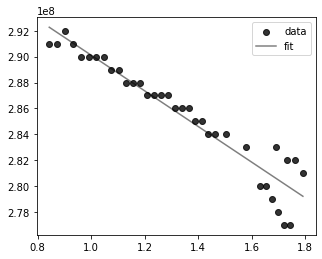
I use the following code to fit a 2 degree polynomial to my 2D data:
deg = 1
fig, ax = plt.subplots(1,facecolor="white")
fig.set_size_inches(5,4)
x = [0.843291,0.873922,0.903581,0.93344,0.961478,0.990968,
1.019434,1.047562,1.075373,1.102883,1.130106,1.157059,
1.183751,1.210195,1.236399,1.262376,1.288134,1.313679,
1.339022,1.364167,1.389123,1.413896,1.438492,1.462916,
1.505685,1.57998,1.690303,1.730423,1.761613,1.791332,
1.63034,1.652749,1.675848,1.698821,1.721672,1.744402]
y = [291000000,291000000,292000000,291000000,290000000,290000000,
290000000,290000000,289000000,289000000,288000000,288000000,
288000000,287000000,287000000,287000000,287000000,286000000,
286000000,286000000,285000000,285000000,284000000,284000000,
284000000,283000000,283000000,282000000,282000000,281000000,
280000000,280000000,279000000,278000000,277000000,277000000]
ax.scatter(x, y,label = "data", c="black", alpha = 0.8)
poly =np.poly1d(np.polyfit(x, y, deg = deg))
ax.plot(np.polyval(poly, x), label='fit', c="black", alpha = 0.5)
ax.legend()
This gives me the following plot, which does not seem realistic:
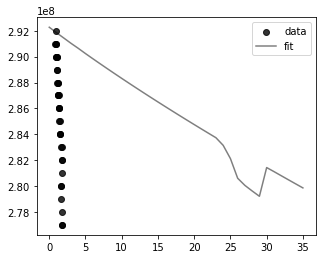
Is there any part of the code I am implementing wrong?
CodePudding user response:
You fit is (almost) correct, your plot isn't:
# first sort the values by x
order = np.argsort(x)
x = np.array(x)[order]
y = np.array(y)[order]
# the plot and compute the fit
deg = 1
fig, ax = plt.subplots(1,facecolor="white")
fig.set_size_inches(5,4)
ax.scatter(x, y,label = "data", c="black", alpha = 0.8)
poly =np.poly1d(np.polyfit(x, y, deg = deg))
# you need to pass the x values here
ax.plot(x, np.polyval(poly, x), label='fit', c="black", alpha = 0.5)
ax.legend()
If you omit the x values, a range is used.
Output: