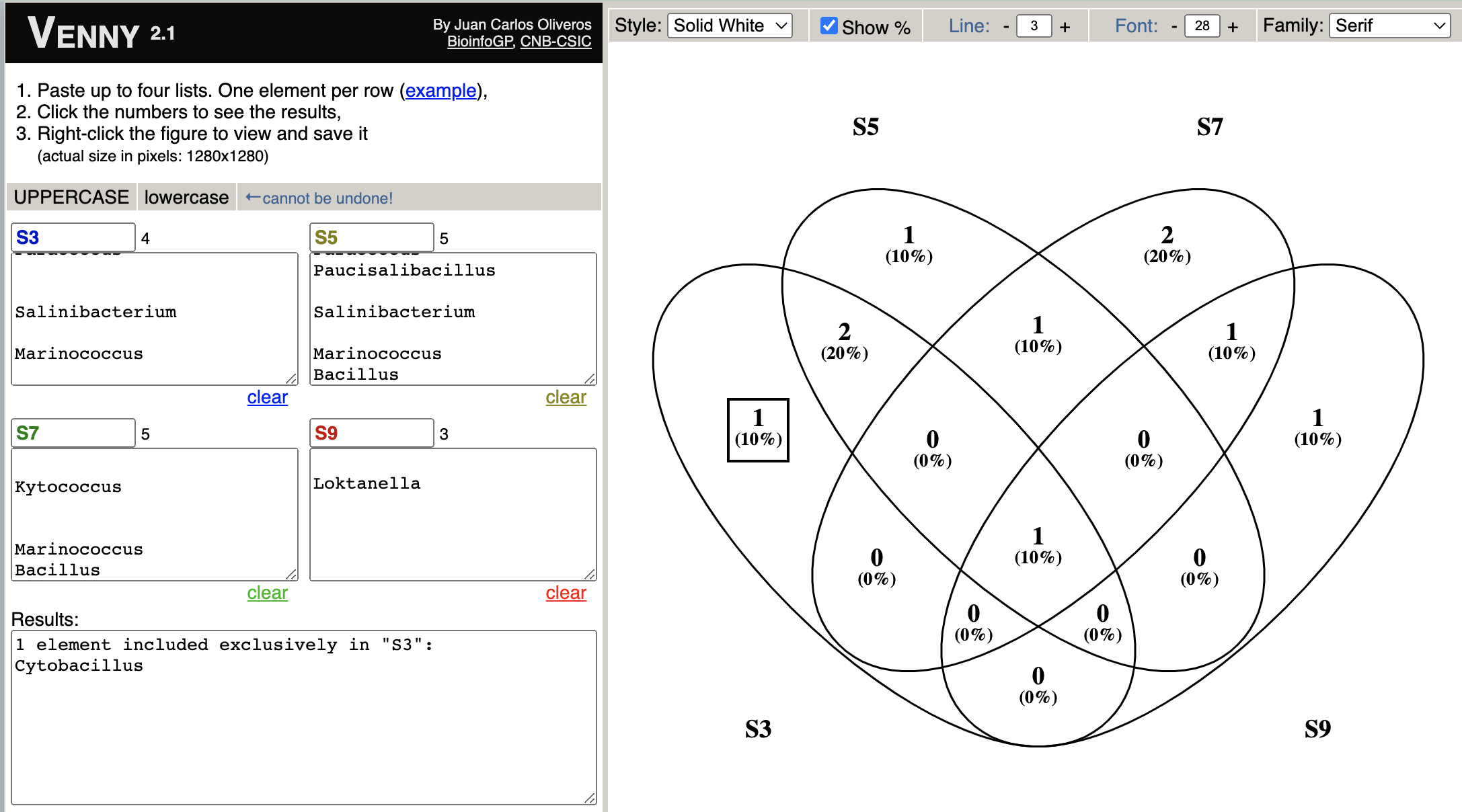
I have a data like contingency table, which display abundance of data, but I want to draw venn diagram from this data fram.
Structure of my data:
species_abundance<-data.frame(Genus = c("Parasphingorhabdus", "Loktanella", "Cytobacillus", "Paracoccus", "Paucisalibacillus", "Kytococcus", "Salinibacterium", "Acinetobacter baumanni","Marinococcus","Bacillus"),
S3 = c(0, 0, 1, 1, 0, 0, 1,0,4,0),
S5 = c(0, 0, 0, 1, 1, 0, 1,0,3,5),
S7 = c(3, 1, 0, 2, 0, 1, 0,0,3,1),
S9 = c(0, 1, 0, 3, 0, 0, 0,1,2,0)
How can I draw venn diagramm from this data frame in order to find unique and shared species accross different stations (S3, S5, S7.......)?
If I convert the data as given below and I try using Venny2 I will get such image, similar image and finding I want to do using R, kindly help
species_abundance1<-data.frame(S3 = c("", "", "Cytobacillus", "Paracoccus", "", "", "Salinibacterium","", "Marinococcus", ""),
S5 = c("", "", "", "Paracoccus", "Paucisalibacillus", "", "Salinibacterium","", "Marinococcus","Bacillus"),
S7 = c("Parasphingorhabdus", "Loktanella", "", "", "", "Kytococcus", "","", "Marinococcus","Bacillus"),
S9 = c("", "Loktanella", "", "", "", "", "","Acinetobacter baumanni", "Marinococcus",""))
CodePudding user response:
There are a couple of ways of getting a 4-variable Venn diagram in R, but Venn diagrams beyond this number of categories are 
A 7-category Venn can't even be drawn using ellipses, and involves a complex floral shape as can be seen in the linked article.
In any case, you can see that a Venn even with 5 categories would not be a very user-friendly way to represent the data.
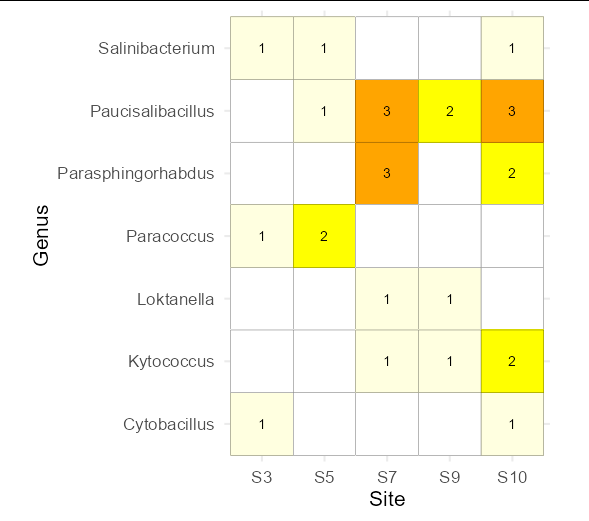
In your case, the natural way to present this kind of data would be via a heatmap. You first need to reshape your data into long format.
library(tidyverse)
species_abundance %>%
pivot_longer(-Genus, names_to = 'Site', values_to = 'Count') %>%
mutate(Site = factor(Site, unique(Site))) %>%
ggplot(aes(Site, Genus, fill = factor(Count)))
geom_tile(color = 'black')
geom_text(aes(label = ifelse(Count == 0, '', Count)))
coord_equal()
scale_fill_manual(guide = 'none',
values = c('white', 'lightyellow', 'yellow', 'orange'))
theme_minimal(base_size = 16)
Addendum
If you really want a 5-category Venn diagram which shows the number of species that the 5 sites have in common, you can do:
library(VennDiagram)
grid::grid.newpage()
with(sign(species_abundance[-1]),
draw.quintuple.venn(sum(S3), sum(S5), sum(S7), sum(S9), sum(S10),
sum(S3 == 1 & S5 == 1), sum(S3 == 1 & S7 == 1),
sum(S3 == 1 & S9 == 1), sum(S3 == 1 & S10 == 1),
sum(S5 == 1 & S7 == 1), sum(S5 == 1 & S9 == 1),
sum(S5 == 1 & S10 == 1), sum(S7 == 1 & S9 == 1),
sum(S7 == 1 & S10 == 1), sum(S9 == 1 & S10 == 1),
sum(S3 == 1 & S5 == 1 & S7 == 1),
sum(S3 == 1 & S5 == 1 & S9 == 1),
sum(S3 == 1 & S5 == 1 & S10 == 1),
sum(S3 == 1 & S7 == 1 & S9 == 1),
sum(S3 == 1 & S7 == 1 & S10 == 1),
sum(S3 == 1 & S9 == 1 & S10 == 1),
sum(S5 == 1 & S7 == 1 & S9 == 1),
sum(S5 == 1 & S7 == 1 & S10 == 1),
sum(S5 == 1 & S9 == 1 & S10 == 1),
sum(S7 == 1 & S9 == 1 & S10 == 1),
sum(S3 == 1 & S5 == 1 & S7 == 1 & S9 == 1),
sum(S3 == 1 & S5 == 1 & S7 == 1 & S10 == 1),
sum(S3 == 1 & S5 == 1 & S9 == 1 & S10 == 1),
sum(S3 == 1 & S7 == 1 & S9 == 1 & S10 == 1),
sum(S5 == 1 & S7 == 1 & S9 == 1 & S10 == 1),
sum(S3 == 1 & S5 == 1 & S7 == 1 & S9 == 1 & S10 == 1),
category = c("S3", "S5", "S7", "S9", "S10"),
fill = c("orange", "red", "green", "blue", "yellow"),
cex = 2,
cat.cex = 2,
cat.col = 'black'
))
Despite being far more difficult to read / understand, this also contains less information than the heatmap. For example, I can see from the Venn that there is a single species that only S3 and S5 have in common, but I can see that just as clearly from the heatmap. Furthermore, I can tell you the Genus (it's paracoccus), and how many observations of it there were at each site using the heatmap. You can't do this with a Venn diagram. A Venn is simply the wrong tool for presenting the data you have.
CodePudding user response:
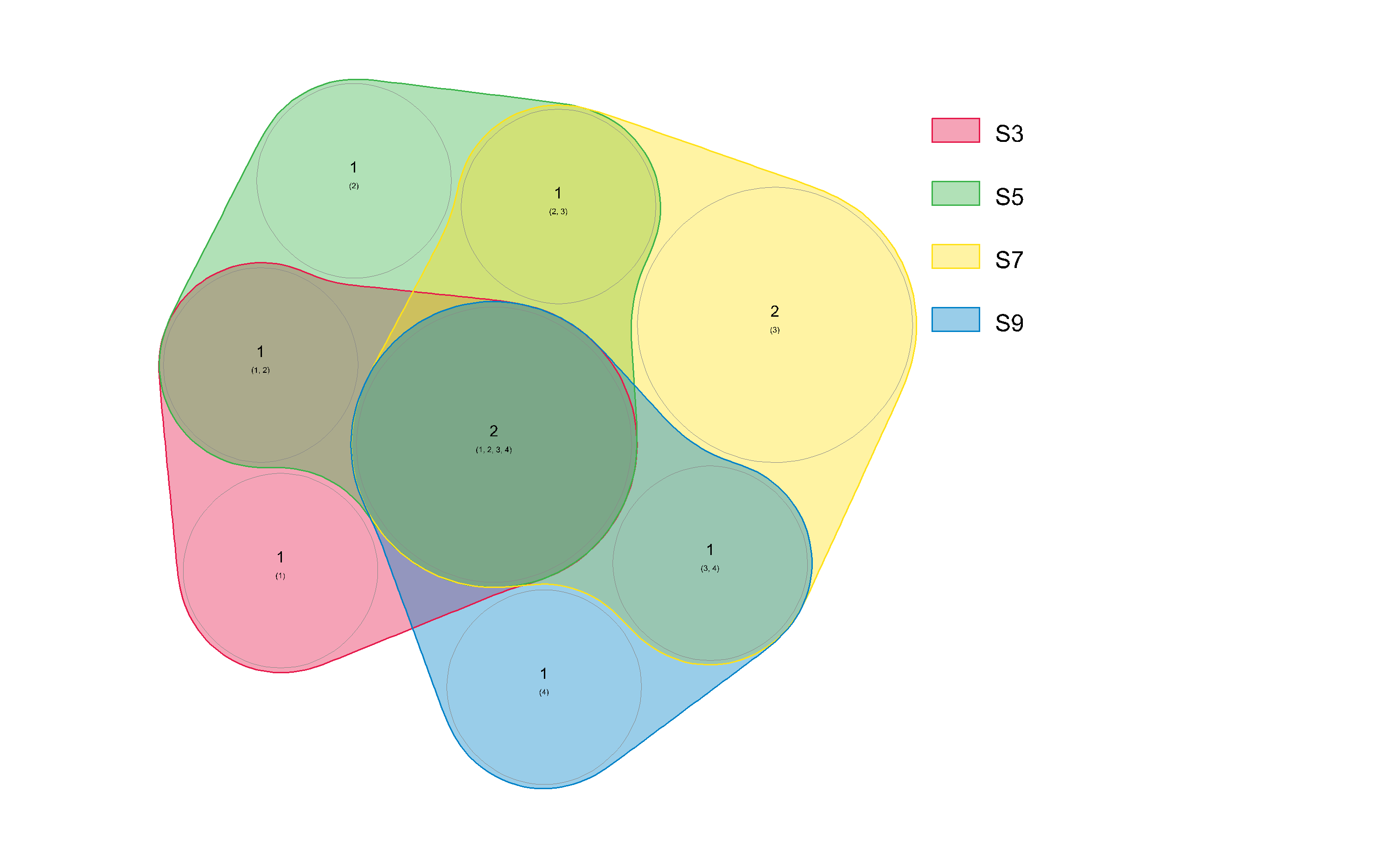
You might consider my nVennR package:
library(nVennR)
species_abundance<-data.frame(Genus = c("Parasphingorhabdus", "Loktanella", "Cytobacillus", "Paracoccus", "Paucisalibacillus", "Kytococcus", "Salinibacterium", "Acinetobacter baumanni","Marinococcus","Bacillus"),
S3 = c(0, 0, 1, 1, 0, 0, 1,0,4,0),
S5 = c(0, 0, 0, 1, 1, 0, 1,0,3,5),
S7 = c(3, 1, 0, 2, 0, 1, 0,0,3,1),
S9 = c(0, 1, 0, 3, 0, 0, 0,1,2,0))
ct <- colnames(species_abundance)
ct <- ct[-1]
r <- vector("list", length = length(ct))
names(r) <- ct
for (v in ct){
t <- species_abundance[species_abundance[,v] != 0, ]$Genus
r[[v]] <- t
}
myV <- plotVenn(r)
You can also browse the results, as shown in the vignette:
> getVennRegion(myV, c("S7"))
[1] "Parasphingorhabdus" "Kytococcus"
> getVennRegion(myV, c("S3", "S7", "S9", "S5"))
[1] "Paracoccus" "Marinococcus"