To be more clear, I produced a 2-D occupancy map, where obstacles marked with white pixels, from 3-D depth data using Python (NumPy) as given in the following figure.
Occupancy Map

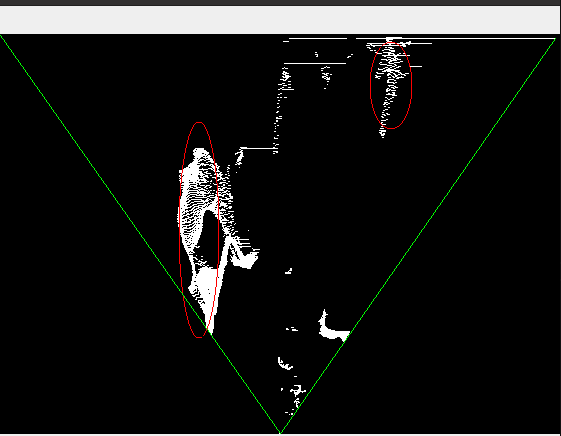
However, occurrence of the objects that are located behind the detected obstacles are also marked but I don't want them to be marked since an obstacle already exist at the front. The following figure shows the case.
Occupancy Map, red ellipses are wrongly marked objects, green lines represent angle of vision
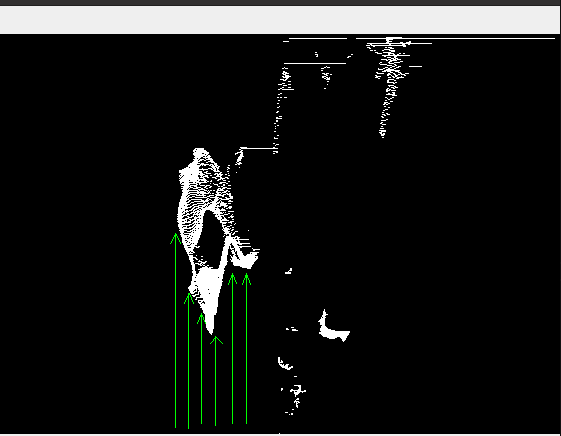
If I simply iterate over the pixels and search for the first occurrence of an obstacle and clear the marks that are located behind it, it would be very slow for real time applications in Python. The iteration concept is given in the following.
Start from left, search for an obstacle from down to up
Also, for some filtering/optimization purposes on depth data, I want to check all the neighbors (8-pixels) of a pixel but not know how to do it with NumPy or any other library. Again, doing with for loops is cumbersome.
Thanks in advance,
Yavuz Selim
'Manual iteration with loops' concept. Fast calculations are expected but not achieved.
CodePudding user response:
I can't see without a for loop in some place, but you can use itertools.groupby:
from itertools import groupby
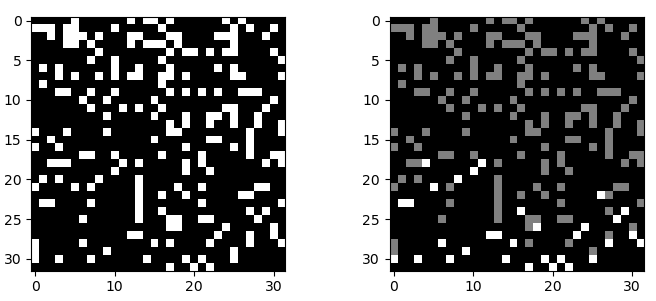
import numpy as np
# Generate data
np.random.seed(34)
data = np.random.choice([0, 1], p=[0.8, 0.2], size=(32, 32))
# Rotate the array because we want to search from "bottom"
# and we will use the fact that np.where sort indices on axis 0
# but axis 0 is the one on the "left"
result = data.copy()
result = np.rot90(result, k=-1)
# Get indices of obstacle
idxs_true = np.where(result == 1)
# Get only one pair for each value column
idxs = [next(grp) for _, grp in groupby(zip(*idxs_true), key=lambda x: x[0])]
idxs = tuple(zip(*idxs))
# Hihglight them
result[idxs] = 2
# Come back to orignal orientation
result = np.rot90(result, k=1)
# Plot
fig, (ax1, ax2) = plt.subplots(ncols=2)
ax1.imshow(data, cmap="gray")
ax2.imshow(result, cmap="gray")
plt.show()