Still quite new to this and am struggling.
I have a directory of a few hundred text files, each file has thousands of lines of information on it.
Some lines contain one number, some many
example:
39 312.000000 168.871795
100.835446
101.800298
102.414406
104.491999
108.855079
107.384008
103.608815
I need to pull all of the information from each text file, I want the name of the text file (minus the '.txt') to be in the first column, and all other information following that to complete the row (regardless of its layout within the file)
import pandas as pd
import os
data= '/path/to/data/'
path='/other/directory/path/'
lst=['list of files needed']
for dirpath, dirs, subj in os.walk(data):
while i<=5: #currently being used to break before iterating through entire directory to check it's working
with open(dirpath lst[i], 'r') as file:
info=file.read().replace('\n', '') #txt file onto one line
corpus.append(lst[i] ' ') #begin list with txt file name
corpus.append(info) #add file contents to list after file name
output=''.join(corpus) #get out of list format
output.split()
i =1
df=pd.read_table(output, lineterminator=',')
df.to_csv(path 'testing.csv')
if i >5:
break
Currently, this is printing Errno 2 (no such file or directory) then goes on to print the contents of the first file and no others, but does not save it to csv.
This also seems horribly convoluted and I'm sure there's another way of doing it
I also suspect the lineterminator will not force each new text file onto a new row, so any suggestions there would be appreciated
desired output:
file1 39 312.000 168.871
file2 72 317.212 173.526
CodePudding user response:
You are loading os and pandas so you can take advantage of their functionality (listdir, path, DataFrame, concat, and to_csv) and drastically reduce your code's complexity.
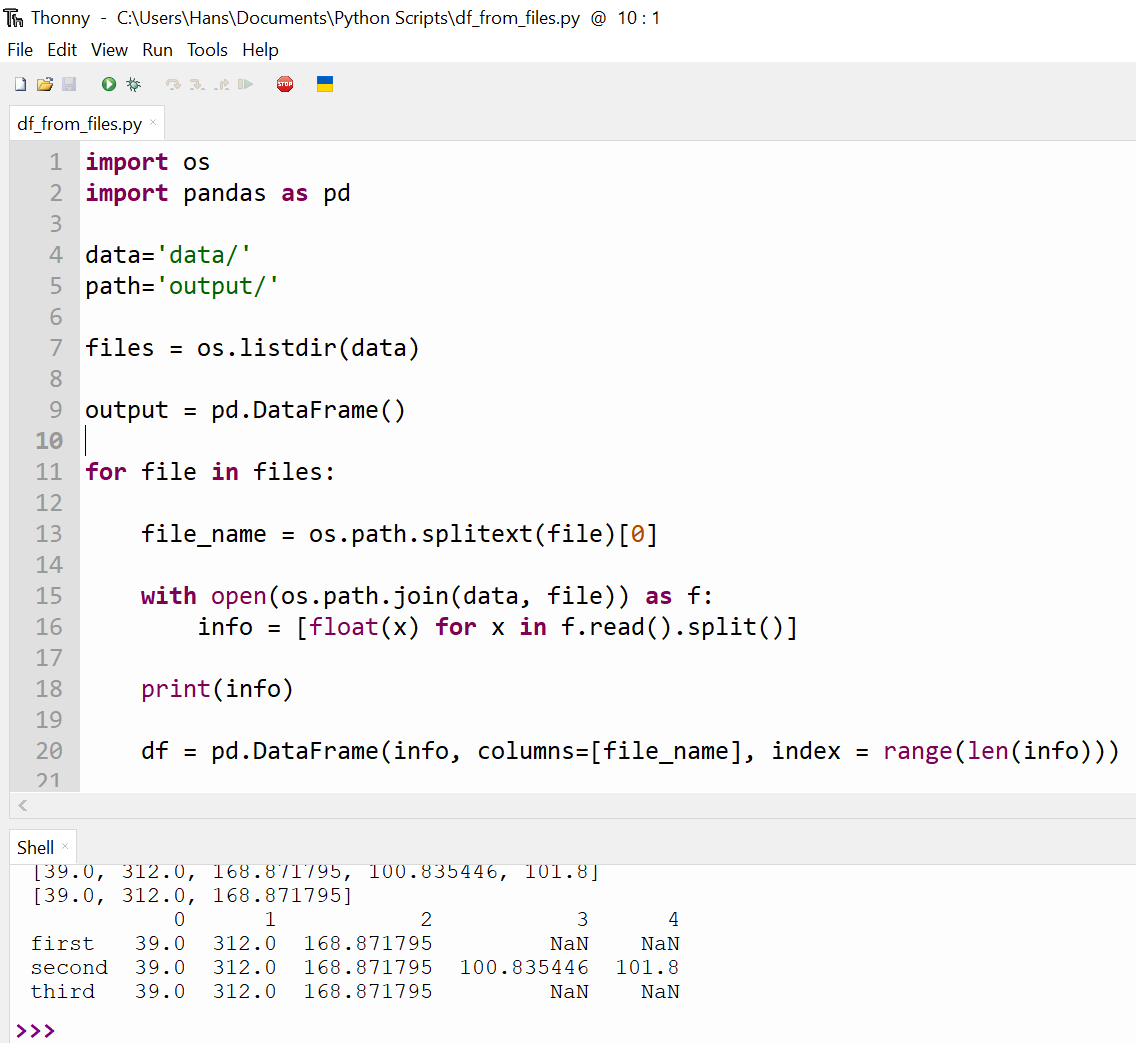
import os
import pandas as pd
data='data/'
path='output/'
files = os.listdir(data)
output = pd.DataFrame()
for file in files:
file_name = os.path.splitext(file)[0]
with open(os.path.join(data, file)) as f:
info = [float(x) for x in f.read().split()]
%print(info)
df = pd.DataFrame(info, columns=[file_name], index = range(len(info)))
output = pd.concat([output, df], axis=1)
output = output.T
print(output)
output.to_csv(path 'testing.csv', index=False)
I would double-check that your data folder only has txt files. And, maybe add a check for txt files to the code.
This got less elegant as I learned about the requirements. If you want to flip the columns and rows, just take out the output.T line. This transposes the dataframe.