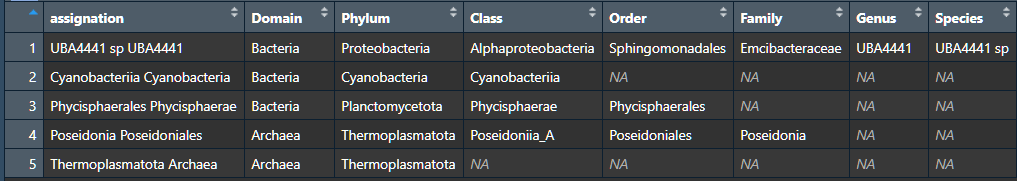
I have a data frame that a portion of it looks like this:
Domain <- c(rep("Bacteria",3),rep("Archaea", 2))
Phylum <- c("Proteobacteria","Cyanobacteria","Planctomycetota", "Thermoplasmatota", "Thermoplasmatota")
Class <- c("Alphaproteobacteria","Cyanobacteriia","Phycisphaerae","Poseidoniia_A",NA)
Order <- c("Sphingomonadales", NA, "Phycisphaerales", "Poseidoniales", NA)
Family <- c("Emcibacteraceae", NA, NA, "Poseidonia", NA)
Genus <- c("UBA4441", NA,NA,NA,NA)
Species <- c("UBA4441 sp", NA,NA,NA,NA)
demo_table <- data.frame(Domain, Phylum, Class, Order, Family, Genus, Species)
The point here is I want to create a new column called "assignation" that consist in the merge of the last two columns that contain non NA values row by row and that the values are separated by a space.
This is the expected output:
| Domain | Phylum | Class | Order | Family | Genus | Species | assignation |
|---|---|---|---|---|---|---|---|
| Bacteria | Proteobacteria | Alphaproteobacteria | Sphingomonadales | Emcibacteraceae | UBA4441 | UBA4441 sp | UBA4441 UBA4441 sp |
| Bacteria | Cyanobacteria | Cyanobacteriia | NA | NA | NA | NA | Cyanobacteria Cyanobacteriia |
| Bacteria | Planctomycetota | Phycisphaerae | Phycisphaerales | NA | NA | NA | Phycisphaerae Phycisphaerales |
| Archaea | Thermoplasmatota | Poseidoniia_A | Poseidoniales | Poseidonia | NA | NA | Poseidoniales Poseidonia |
| Archaea | Thermoplasmatota | NA | NA | NA | NA | NA | Archaea Thermoplasmatota |
I Think that paste() may work on this case but not sure how to implement it so I can get the above mentioned expected output data frame.
CodePudding user response:
We may use base R - loop over the rows, remove the NA with na.omit, get the last two elements tail with n = 2 and paste
demo_table$assignation <- apply(demo_table, 1,
function(x) paste(tail(na.omit(x), 2), collapse = " "))
-output
demo_table$assignation
[1] "UBA4441 UBA4441 sp" "Cyanobacteria Cyanobacteriia" "Phycisphaerae Phycisphaerales" "Poseidoniales Poseidonia"
[5] "Archaea Thermoplasmatota"
With tidyverse, we may also use unite and remove the NA with na.rm = TRUE, then extract the last two words
library(dplyr)
library(tidyr)
library(stringr)
demo_table %>%
unite(assignation, everything(), na.rm = TRUE, remove = FALSE) %>%
mutate(assignation = str_replace(assignation,
".*_([^_] )_([^_] )$", "\\1 \\2")) %>%
relocate(assignation, .after =last_col())
CodePudding user response:
If you want to go for a tidyverse approach, you just need to use rowwise c_across. I think is it also nice to transform this operation in a function, in case you need to use later or even change the behavior of it.
Code
library(dplyr)
select_last <- function(x, n = 2){paste(tail(na.omit(x),n = n),collapse = " ")}
demo_table %>%
rowwise() %>%
mutate(assignation = select_last(c_across()))
Output
# A tibble: 5 x 8
# Rowwise:
Domain Phylum Class Order Family Genus Species assignation
<chr> <chr> <chr> <chr> <chr> <chr> <chr> <chr>
1 Bacter~ Proteobact~ Alphaproteo~ Sphingomon~ Emcibacte~ UBA4~ UBA4441~ UBA4441 UBA4441 sp
2 Bacter~ Cyanobacte~ Cyanobacter~ NA NA NA NA Cyanobacteria Cyan~
3 Bacter~ Planctomyc~ Phycisphaer~ Phycisphae~ NA NA NA Phycisphaerae Phyc~
4 Archaea Thermoplas~ Poseidoniia~ Poseidonia~ Poseidonia NA NA Poseidoniales Pose~
5 Archaea Thermoplas~ NA NA NA NA NA Archaea Thermoplas~
CodePudding user response:
Here is dplyr combined with tidyr approach:
library(dplyr)
library(tidyr)
demo_table %>%
mutate(id = row_number()) %>%
pivot_longer(-id) %>%
group_by(id) %>%
na.omit() %>%
arrange(-row_number(), .by_group = TRUE) %>%
mutate(assignation = paste(value[1], value[2], sep = "\n")) %>%
slice(1) %>%
ungroup() %>%
select(assignation) %>%
bind_cols(demo_table) %>%
View()