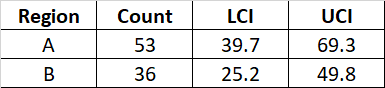
I am attempting to draw out confidence limits from some count data I have. I can do this manually, for example
poisson.test(53,conf.level = 0.95)
LCI = 39.7, UCI = 69.3
So I have some basics! I have some very very basic test data to practice on, but I am having trouble applying poisson.test for each row and then extracting the LCI/UCI. The dataset is:
region <- c("A","B")
count <- c(53,36)
data <- data.frame(region,count)
Ideally, my output would look like this:
I have tried using apply() but that hasn't quite worked. Any tips on how to get this out and into a table without having to perform the manual calculation and enter it into a spreadsheet? Thanks in advance!
CodePudding user response:
Select the count column and apply poisson.test on each of its element with sapply.
data[c("LCI", "UCI")] <- t(sapply(data[["count"]], function(x) poisson.test(x)$conf.int))
# region count LCI UCI
# 1 A 53 39.70064 69.32530
# 2 B 36 25.21396 49.83917
CodePudding user response:
You can use the sapply() function to apply the poisson.test() function to each row of your data frame and extract the lower and upper confidence intervals. Here's an example:
library(stats)
# Add columns for lower and upper confidence intervals
data$LCI <- sapply(data$count, function(x) poisson.test(x, conf.level = 0.95)$conf.int[1])
data$UCI <- sapply(data$count, function(x) poisson.test(x, conf.level = 0.95)$conf.int[2])
# Print the modified data frame
data
This will add two new columns to the data frame, LCI and UCI with the corresponding lower and upper confidence intervals for each count.
Alternatively, you can also use the tidy() function from the broom package to extract the confidence intervals from the poisson.test output and then join it with the data frame, like this:
library(broom)
data_tidy <- data %>%
mutate(poisson = map(count, poisson.test)) %>%
unnest(poisson, .key = "interval", .value = "poisson") %>%
select(region, count, interval)
data_tidy
This will give you a new data frame with columns region, count, interval which contains the poisson.test output, and you can extract the LCI/UCI from interval column by selecting the respective element.