I have a data frame with 25 features as columns, call it df1. In another data frame, call it df2, there are two label columns that I want to use. The first label column is cancertype and it has only three cancer types. The second label is stage and it has 4 different stages. Both df1 and df2 have the same exact samples in the rows, in the same order.
I want to create a boxplot for each feature, where the x-axis would be the three cancer types, and for each cancer type, I'd have 4 box plots according to the stage. So in the end, I'll have 25 boxplots, each one for each feature.
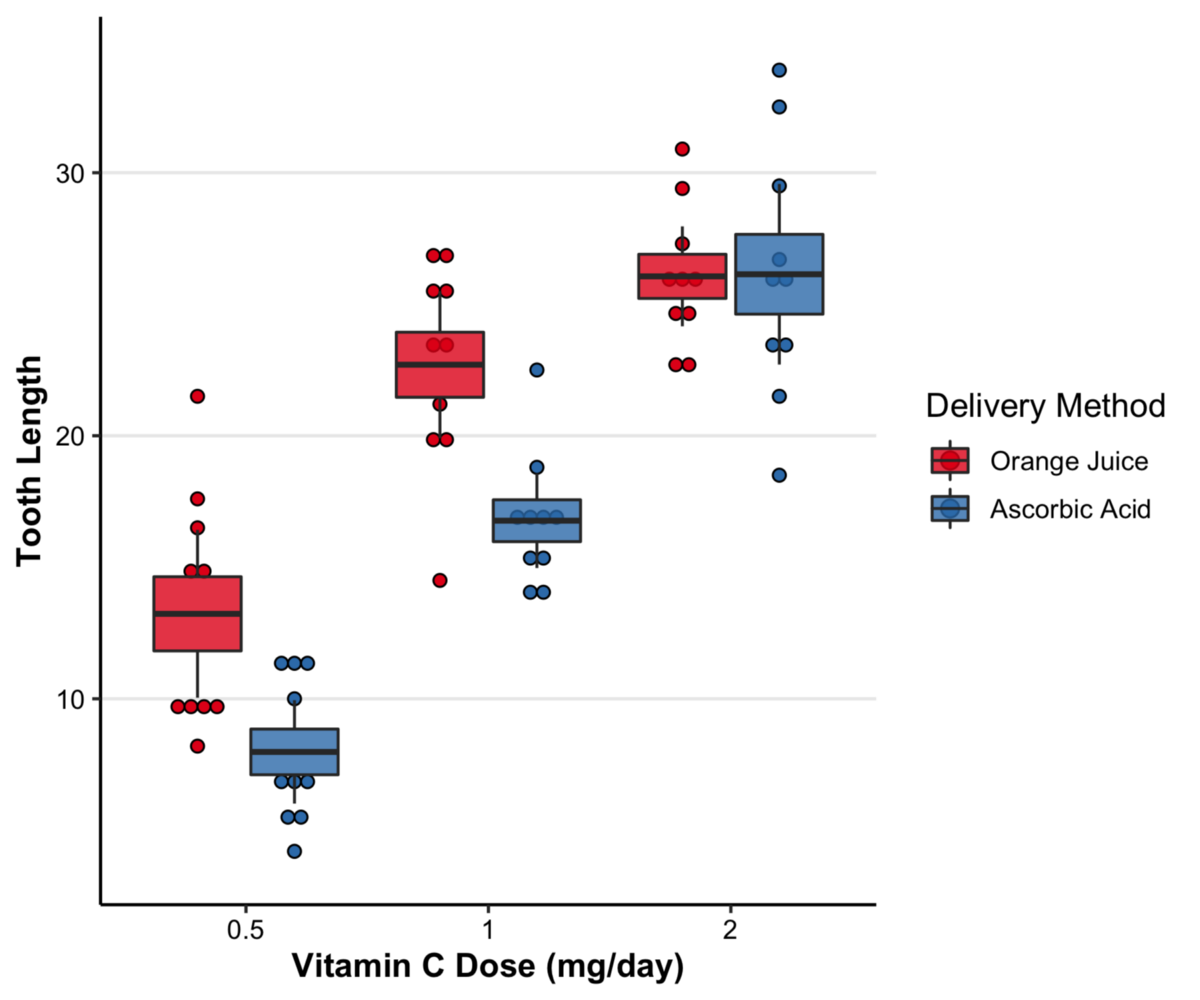
Here is a plot as an example:
So here instead of 0.5, 1, and 2, I'd have the three cancer types, and instead of Orange juice and Ascorbic juice, I'll have the 4 stages of cancer. And of course, instead of Tooth Length, I'll have the values of the feature. How can I do that in R?
Here is an example of the data, I combined the two labels columns stage and cancertype from df2, to some of the features of df1, just so you guys see what the data looks like:
structure(list(`CD4-T-cells` = c(-0.126653261025908, -0.146944662222103,
-0.0115148964617506, -0.215846341670589, -0.146791213061172,
-0.219857803332179, -0.17081620282989, -0.0891711254320417, -0.0874442820512679,
-0.220366749282219, -0.133508075707128, -0.193271241796752, -0.201211825907853,
-0.125225983272556, -0.115043218071495), `CD8-T-cells` = c(-0.157666852910288,
-0.232728699747416, -0.0160616032665578, -0.209410515957277,
-0.202956751240547, -0.175764455863453, -0.323439446327317, -0.0338852795521566,
-0.135834830888521, -0.141126851397699, -0.255009952032843, -0.213879070793134,
-0.183317103095461, -0.189072789833267, -0.259816019334781),
`T-helpers` = c(-0.22607814653843, -0.215166591267584, -0.00118837391747009,
-0.312608050611363, -0.341117207247747, -0.294448125003604,
-0.292393762983215, -0.118961302821571, -0.253841509210292,
-0.183748456919948, -0.260611567417525, -0.265980836082014,
-0.200401095547006, -0.189306997555047, -0.284547076718132
), `NK-cells` = c(-0.0768979654779382, -0.156845903079279,
-0.00175695677961327, 0.0435753727359533, -0.113414879053929,
-0.0719972487895254, -0.173138898156026, -0.0363772958157232,
-0.113696954024299, -0.0452152425828402, -0.169035653930283,
-0.0850518565032839, -0.0992324194948109, -0.0820755296254414,
-0.158947253410155), cancertype = c("Melanoma", "Urothelial Bladder Carcinoma",
"Urothelial Bladder Carcinoma", "Renal Clear Cell Carcinoma",
"Renal Clear Cell Carcinoma", "Renal Clear Cell Carcinoma",
"Renal Clear Cell Carcinoma", "Melanoma", "Renal Clear Cell Carcinoma",
"Melanoma", "Renal Clear Cell Carcinoma", "Melanoma", "Melanoma",
"Renal Clear Cell Carcinoma", "Urothelial Bladder Carcinoma"
), stage = c("stage1", "stage2", "stage4", "Stage3", " Stage2",
"Stage4", "Stage1", "Stage1", "Stage1", " Stage1", "Stage3",
"Stage1", "Stage3", "Stage3", "Stage4")), class = "data.frame", row.names = c("Pt1",
"Pt10", "Pt101", "Pt103", "Pt106", "Pt11", "Pt17", "Pt18", "Pt2",
"Pt24", "Pt26", "Pt27", "Pt28", "Pt29", "Pt3"))
EDIT -
Apparently, I have too many features so presenting the plots together with facet_wrap would make a mess. Therefore, what I need is to save each plot individually inside a list, and then for example, I call mylist[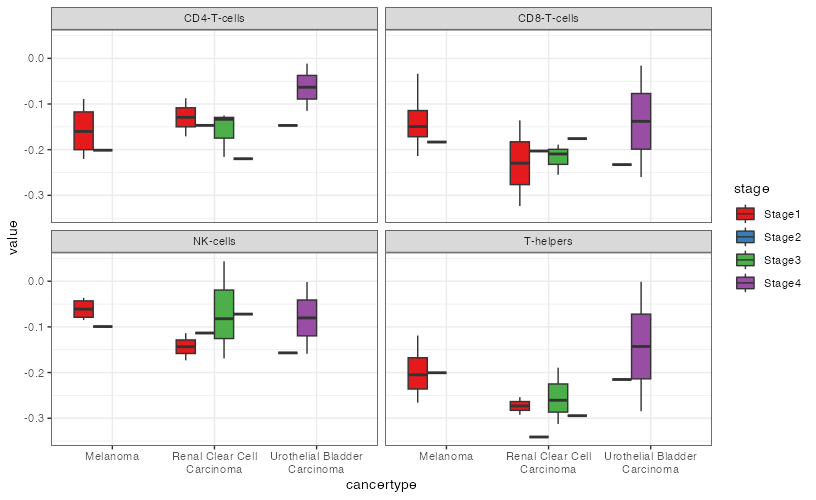
EDIT And to output one plot per feature as a list you could do:
library(tidyverse)
dat |>
pivot_longer(-c(cancertype, stage), names_to = "feature") |>
mutate(stage = stringr::str_to_title(stage),
stage = stringr::str_trim(stage)) |>
group_split(feature) |>
lapply(function(.data) {
ggplot(.data, aes(cancertype, value, fill = stage))
geom_boxplot(position = position_dodge(preserve = "single"))
scale_x_discrete(labels = ~str_wrap(.x, 25))
scale_fill_brewer(palette = "Set1")
labs(title = unique(.data$feature))
theme_bw()
})