Let's say I have two DataFrames (Samples and Controls) as follows:
df_Sample =\
{'Nuclei in individual cell region Selected - Nucleus Area [µm²]': {0: 189.48, 1: 153.736, 2: 199.219, 3: 221.4, 4: 261.648, 5: 304.089, 6: 345.935, 7: 218.935, 8: 232.601, 9: 240.912, 10: 208.125, 11: 260.713, 12: 161.112, 13: 270.181, 14: 165.888, 15: 342.077, 16: 158.376, 17: 557.035, 18: 319.913, 19: 257.297},
'Nuclei in individual cell region Selected - Nucleus Roundness': {0: 0.913951, 1: 0.93739, 2: 0.93725, 3: 0.869216, 4: 0.828391, 5: 0.978106, 6: 0.955958, 7: 0.92616, 8: 0.78398, 9: 0.977184, 10: 0.848469, 11: 0.984681, 12: 0.908689, 13: 0.910773, 14: 0.908787, 15: 0.986723, 16: 0.976819, 17: 0.95381, 18: 0.976402, 19: 0.930968},
'Nuclei in individual cell region Selected - Nucleus Width [µm]': {0: 11.4282, 1: 12.2188, 2: 13.9467, 3: 12.9901, 4: 14.3977, 5: 17.4717, 6: 17.0762, 7: 14.3598, 8: 11.9658, 9: 15.5159, 10: 14.1908, 11: 15.9906, 12: 11.1176, 13: 15.854, 14: 12.266, 15: 18.1792, 16: 12.6883, 17: 22.2749, 18: 18.5788, 19: 14.8166},
'Nuclei in individual cell region Selected - Nucleus Length [µm]': {0: 18.9918, 1: 15.8738, 2: 16.5248, 3: 19.1131, 4: 21.3145, 5: 20.084, 6: 24.1163, 7: 18.2035, 8: 22.8184, 9: 19.0128, 10: 18.5242, 11: 21.1097, 12: 16.8669, 13: 21.2989, 14: 16.8885, 15: 23.6588, 16: 15.8094, 17: 29.3571, 18: 21.1347, 19: 19.8769},
'Nuclei in individual cell region Selected - Nucleus Ratio Width to Length': {0: 0.601743, 1: 0.769748, 2: 0.843986, 3: 0.679645, 4: 0.675488, 5: 0.869933, 6: 0.708077, 7: 0.788848, 8: 0.524394, 9: 0.816074, 10: 0.766064, 11: 0.757499, 12: 0.659136, 13: 0.744356, 14: 0.726293, 15: 0.768394, 16: 0.80258, 17: 0.758756, 18: 0.879065, 19: 0.745417},
'Nuclei in individual cell region Selected - Nucleus HOECHST 33342 Haralick Correlation 1 px': {0: 0.98371, 1: 0.97789, 2: 0.978729, 3: 0.961711, 4: 0.976911, 5: 0.966404, 6: 0.98986, 7: 0.972134, 8: 0.970894, 9: 0.949579, 10: 0.964805, 11: 0.970876, 12: 0.966332, 13: 0.978358, 14: 0.984657, 15: 0.965988, 16: 0.989449, 17: 0.970398, 18: 0.962764, 19: 0.962354},
'Nuclei in individual cell region Selected - Nucleus HOECHST 33342 Haralick Contrast 1 px': {0: 0.00262663, 1: 0.00337056, 2: 0.00384226, 3: 0.00407926, 4: 0.00339842, 5: 0.00268196, 6: 0.00258363, 7: 0.0026726, 8: 0.0039011, 9: 0.0049614, 10: 0.00584036, 11: 0.00359065, 12: 0.00503498, 13: 0.00360473, 14: 0.00342672, 15: 0.00324812, 16: 0.00266534, 17: 0.00354377, 18: 0.00508052, 19: 0.00399667},
'Nuclei in individual cell region Selected - Nucleus HOECHST 33342 Haralick Sum Variance 1 px': {0: 0.0799574, 1: 0.075373, 2: 0.089302, 3: 0.0522426, 4: 0.0727336, 5: 0.0392431, 6: 0.12669, 7: 0.0472695, 8: 0.0660276, 9: 0.0479593, 10: 0.0815123, 11: 0.0607464, 12: 0.0735158, 13: 0.0823799, 14: 0.110817, 15: 0.0469307, 16: 0.125631, 17: 0.0589657, 18: 0.0669395, 19: 0.0520771},
'Nuclei in individual cell region Selected - Nucleus HOECHST 33342 Haralick Homogeneity 1 px': {0: 0.739913, 1: 0.68523, 2: 0.695601, 3: 0.671093, 4: 0.708442, 5: 0.753666, 6: 0.787906, 7: 0.727063, 8: 0.680108, 9: 0.634683, 10: 0.626611, 11: 0.687146, 12: 0.661779, 13: 0.678676, 14: 0.695092, 15: 0.724737, 16: 0.748956, 17: 0.697572, 18: 0.647701, 19: 0.677194},
'Nuclei in individual cell region Selected - Nucleus HOECHST 33342 SER Spot 0 px': {0: 0.005843, 1: 0.00580018, 2: 0.0071962, 3: 0.00964391, 4: 0.00578204, 5: 0.00631538, 6: 0.00591882, 7: 0.00738057, 8: 0.00797945, 9: 0.0107222, 10: 0.00789028, 11: 0.0079751, 12: 0.00720769, 13: 0.00583212, 14: 0.00612275, 15: 0.00729683, 16: 0.00605783, 17: 0.00678319, 18: 0.00903149, 19: 0.00873706},
'Nuclei in individual cell region Selected - Nucleus HOECHST 33342 SER Hole 0 px': {0: 0.0053161, 1: 0.00527502, 2: 0.00624592, 3: 0.00904184, 4: 0.00543591, 5: 0.00533345, 6: 0.00579994, 7: 0.00647572, 8: 0.00731868, 9: 0.0104302, 10: 0.00760632, 11: 0.00771892, 12: 0.00689596, 13: 0.00578755, 14: 0.00604904, 15: 0.00727409, 16: 0.00561067, 17: 0.00706209, 18: 0.00924693, 19: 0.00861305},
'Nuclei in individual cell region Selected - Nucleus HOECHST 33342 SER Edge 0 px': {0: 0.0554048, 1: 0.0704348, 2: 0.062886, 3: 0.0676434, 4: 0.0616821, 5: 0.0566622, 6: 0.0475497, 7: 0.056854, 8: 0.0712491, 9: 0.077949, 10: 0.0817617, 11: 0.0688477, 12: 0.0827153, 13: 0.0629512, 14: 0.0608878, 15: 0.0607465, 16: 0.0560636, 17: 0.0645136, 18: 0.0726108, 19: 0.066896},
'Nuclei in individual cell region Selected - Nucleus HOECHST 33342 SER Ridge 0 px': {0: 0.00924915, 1: 0.00908236, 2: 0.0118103, 3: 0.0165759, 4: 0.0101151, 5: 0.0109813, 6: 0.00959717, 7: 0.0121257, 8: 0.0136556, 9: 0.0180968, 10: 0.0136057, 11: 0.0143802, 12: 0.014296, 13: 0.00956464, 14: 0.0105358, 15: 0.0127249, 16: 0.00991149, 17: 0.012284, 18: 0.015938, 19: 0.0156756},
'Nuclei in individual cell region Selected - Nucleus HOECHST 33342 SER Valley 0 px': {0: 0.0104073, 1: 0.0108218, 2: 0.0132724, 3: 0.0186756, 4: 0.012417, 5: 0.0120152, 6: 0.0107475, 7: 0.0132826, 8: 0.0163031, 9: 0.0216996, 10: 0.0181437, 11: 0.0155132, 12: 0.018504, 13: 0.0125872, 14: 0.012248, 15: 0.0145793, 16: 0.0104176, 17: 0.0148176, 18: 0.0189796, 19: 0.0183744},
'Nuclei in individual cell region Selected - Nucleus HOECHST 33342 SER Saddle 0 px': {0: 0.0110422, 1: 0.0115229, 2: 0.0137925, 3: 0.0184715, 4: 0.012461, 5: 0.0114347, 6: 0.00987503, 7: 0.0135181, 8: 0.0158798, 9: 0.0205525, 10: 0.017767, 11: 0.0154586, 12: 0.0151242, 13: 0.0124683, 14: 0.0119072, 15: 0.0141378, 16: 0.0104225, 17: 0.0142464, 18: 0.0184273, 19: 0.0172968},
'Nuclei in individual cell region Selected - Nucleus HOECHST 33342 SER Bright 0 px': {0: 0.0131424, 1: 0.012963, 2: 0.0165551, 3: 0.0228766, 4: 0.0138591, 5: 0.0150853, 6: 0.0135239, 7: 0.0169965, 8: 0.0188593, 9: 0.0251123, 10: 0.0187394, 11: 0.0194767, 12: 0.01881, 13: 0.013414, 14: 0.0145416, 15: 0.0174515, 16: 0.0138995, 17: 0.0166307, 18: 0.0217725, 19: 0.0213088},
'Nuclei in individual cell region Selected - Nucleus HOECHST 33342 SER Dark 0 px': {0: 0.0137252, 1: 0.0140704, 2: 0.017077, 3: 0.0242349, 4: 0.0156365, 5: 0.0152167, 6: 0.0145082, 7: 0.0172853, 8: 0.0206896, 9: 0.0281842, 10: 0.0225596, 11: 0.0203449, 12: 0.0224352, 13: 0.016074, 14: 0.0160069, 15: 0.0191488, 16: 0.0139954, 17: 0.0191773, 18: 0.0247077, 19: 0.0236879},
'Nuclei in individual cell region Selected - Intensity Nucleus HOECHST 33342 Mean': {0: 10439.2, 1: 8599.48, 2: 11024.7, 3: 14120.2, 4: 13009.2, 5: 14328.9, 6: 8880.34, 7: 13258.0, 8: 13797.4, 9: 11089.1, 10: 8444.29, 11: 18060.7, 12: 12378.4, 13: 10022.7, 14: 11975.5, 15: 10022.7, 16: 7041.5, 17: 13130.3, 18: 16532.3, 19: 13920.7},
'Nuclei in individual cell region Selected - Intensity Nucleus HOECHST 33342 StdDev': {0: 3146.52, 1: 2589.1, 2: 3462.54, 3: 3468.93, 4: 3741.13, 5: 3113.11, 6: 3266.78, 7: 3160.88, 8: 3893.39, 9: 2664.13, 10: 2586.55, 11: 4766.58, 12: 3712.11, 13: 3047.99, 14: 4211.4, 15: 2354.91, 16: 2635.87, 17: 3371.18, 18: 4531.04, 19: 3411.83},
'Nuclei in individual cell region Selected - Individual Cell Region resized Area [µm²]': {0: 445.553, 1: 397.35, 2: 442.885, 3: 510.77, 4: 697.139, 5: 915.99, 6: 1016.63, 7: 528.905, 8: 778.639, 9: 729.705, 10: 611.068, 11: 532.118, 12: 413.038, 13: 951.751, 14: 316.65, 15: 1195.33, 16: 490.731, 17: 1677.82, 18: 1153.86, 19: 769.885},
'Nuclei in individual cell region Selected - Individual Cell Region resized Roundness': {0: 0.857263, 1: 0.795805, 2: 0.814236, 3: 0.854813, 4: 0.831398, 5: 0.777984, 6: 0.787167, 7: 0.747858, 8: 0.750062, 9: 0.762677, 10: 0.771427, 11: 0.780667, 12: 0.884383, 13: 0.666342, 14: 0.765064, 15: 0.808236, 16: 0.85367, 17: 0.79878, 18: 0.630026, 19: 0.838658},
'Nuclei in individual cell region Selected - Individual Cell Region resized Width [µm]': {0: 20.4397, 1: 18.2035, 2: 17.217, 3: 18.6955, 4: 22.8935, 5: 24.9457, 6: 27.1186, 7: 19.1837, 8: 20.5044, 9: 24.3093, 10: 19.5575, 11: 21.0186, 12: 17.3154, 13: 23.012, 14: 16.2186, 15: 26.8312, 16: 21.4016, 17: 32.6773, 18: 27.1085, 19: 25.9816},
'Nuclei in individual cell region Selected - Individual Cell Region resized Length [µm]': {0: 28.0335, 1: 28.1183, 2: 31.5599, 3: 31.9347, 4: 36.3173, 5: 51.6394, 6: 41.2543, 7: 38.9602, 8: 52.7941, 9: 43.4318, 10: 42.1264, 11: 36.0593, 12: 30.6021, 13: 50.7546, 14: 24.1592, 15: 56.6319, 16: 27.9525, 17: 61.0174, 18: 57.4963, 19: 42.2456},
'Nuclei in individual cell region Selected - Individual Cell Region resized Ratio Width to Length': {0: 0.729115, 1: 0.647391, 2: 0.545533, 3: 0.585429, 4: 0.630374, 5: 0.483074, 6: 0.65735, 7: 0.492392, 8: 0.388385, 9: 0.559713, 10: 0.464257, 11: 0.58289, 12: 0.565824, 13: 0.453397, 14: 0.671319, 15: 0.473783, 16: 0.765642, 17: 0.53554, 18: 0.471483, 19: 0.615013},
'Nuclei in individual cell region Selected - Relative Spot Intensity': {0: 0.00431319, 1: 0.0207483, 2: 0.0272823, 3: 0.0526484, 4: 0.0874202, 5: 0.0260405, 6: 0.0325056, 7: 0.0588061, 8: 0.0335587, 9: 0.0496844, 10: 0.0273733, 11: 0.0306711, 12: 0.014466, 13: 0.0147694, 14: 0.0207914, 15: 0.0134007, 16: 0.0534635, 17: 0.0133466, 18: 0.113961, 19: 0.00055431},
'Nuclei in individual cell region Selected - Number of Spots per Area of Individual Cell Region resized': {0: 0.000228885, 1: 0.000299427, 2: 0.000460529, 3: 0.000898473, 4: 0.00112151, 5: 0.000575225, 6: 0.000618595, 7: 0.00144611, 8: 0.000720351, 9: 0.000163049, 10: 0.000361593, 11: 0.000511068, 12: 0.000329205, 13: 0.000375027, 14: 0.000536769, 15: 0.000270167, 16: 0.000831255, 17: 0.000344429, 18: 0.00138465, 19: 2.2077e-05},
'Compound': {0: 'Ciprofloxacin-Low', 1: 'Flunisolide-Medium', 2: 'Famprofazone-Medium', 3: 'Alprenolol-High', 4: 'Dyclonine-Low', 5: 'Flunisolide-Medium', 6: 'Zaleplon-Medium', 7: 'Hexetidine-Low', 8: 'Hexetidine-High', 9: 'Amprolium-Medium', 10: 'Pindolol-Low', 11: 'Zaleplon-High', 12: 'Famprofazone-Low', 13: 'Dyclonine-High', 14: 'Montensin-Medium', 15: 'Pindolol-Medium', 16: 'Hexetidine-Medium', 17: 'Flunisolide-Medium', 18: 'Dyclonine-Medium', 19: 'Hexetidine-Low'}}
df1_Sample = pd.DataFrame(df_Sample)
df_Control =\
{'Nuclei in individual cell region Selected - Nucleus Area [µm²]': {106695: 205.185, 106696: 160.008, 106697: 329.227, 106698: 264.521, 106699: 242.867, 106700: 225.598, 106701: 53.7438, 106702: 63.8908, 106703: 208.244, 106704: 195.48, 106705: 218.51, 106706: 160.262, 106707: 190.568, 106708: 254.697, 106709: 239.399, 106710: 59.5907, 106711: 228.267, 106712: 164.512, 106713: 125.691, 106714: 177.412},
'Nuclei in individual cell region Selected - Nucleus Roundness': {106695: 0.985695, 106696: 0.679483, 106697: 0.980048, 106698: 0.918674, 106699: 0.882368, 106700: 0.910482, 106701: 0.833087, 106702: 0.915233, 106703: 0.981635, 106704: 0.944526, 106705: 0.949615, 106706: 0.757661, 106707: 0.939818, 106708: 0.950865, 106709: 0.941393, 106710: 0.817561, 106711: 0.919093, 106712: 0.973769, 106713: 0.944191, 106714: 0.956228},
'Nuclei in individual cell region Selected - Nucleus Width [µm]': {106695: 12.7764, 106696: 10.5496, 106697: 18.2818, 106698: 14.348, 106699: 10.9667, 106700: 11.5818, 106701: 5.76001, 106702: 7.3426, 106703: 14.0801, 106704: 12.031, 106705: 13.4403, 106706: 11.6433, 106707: 12.6239, 106708: 13.4706, 106709: 13.9272, 106710: 6.47673, 106711: 12.4858, 106712: 12.6239, 106713: 10.9543, 106714: 12.5293},
'Nuclei in individual cell region Selected - Nucleus Length [µm]': {106695: 19.4166, 106696: 16.8765, 106697: 22.8452, 106698: 23.532, 106699: 24.0351, 106700: 22.2779, 106701: 9.97151, 106702: 10.0935, 106703: 18.1891, 106704: 19.4324, 106705: 19.2288, 106706: 15.9256, 106707: 17.6098, 106708: 24.0853, 106709: 20.7766, 106710: 10.9706, 106711: 19.783, 106712: 15.9821, 106713: 14.4354, 106714: 17.575},
'Nuclei in individual cell region Selected - Nucleus Ratio Width to Length': {106695: 0.658015, 106696: 0.62511, 106697: 0.800247, 106698: 0.609723, 106699: 0.45628, 106700: 0.519879, 106701: 0.577646, 106702: 0.727458, 106703: 0.774099, 106704: 0.61912, 106705: 0.698966, 106706: 0.731104, 106707: 0.716864, 106708: 0.559289, 106709: 0.670332, 106710: 0.590371, 106711: 0.631136, 106712: 0.789875, 106713: 0.758852, 106714: 0.7129},
'Nuclei in individual cell region Selected - Nucleus HOECHST 33342 Haralick Correlation 1 px': {106695: 0.973546, 106696: 0.970546, 106697: 0.967139, 106698: 0.974698, 106699: 0.968529, 106700: 0.972811, 106701: 0.978456, 106702: 0.972309, 106703: 0.975749, 106704: 0.97255, 106705: 0.977455, 106706: 0.965869, 106707: 0.977174, 106708: 0.969181, 106709: 0.977156, 106710: 0.979732, 106711: 0.975186, 106712: 0.97187, 106713: 0.978189, 106714: 0.975682},
'Nuclei in individual cell region Selected - Nucleus HOECHST 33342 Haralick Contrast 1 px': {106695: 0.00425443, 106696: 0.00819948, 106697: 0.00291286, 106698: 0.00296901, 106699: 0.00336917, 106700: 0.00358292, 106701: 0.00548305, 106702: 0.00543524, 106703: 0.00346719, 106704: 0.00445449, 106705: 0.00386494, 106706: 0.00941484, 106707: 0.00300193, 106708: 0.00308412, 106709: 0.00300024, 106710: 0.0049655, 106711: 0.00337084, 106712: 0.00346975, 106713: 0.00513168, 106714: 0.00352557},
'Nuclei in individual cell region Selected - Nucleus HOECHST 33342 Haralick Sum Variance 1 px': {106695: 0.0793487, 106696: 0.137136, 106697: 0.0435847, 106698: 0.0579307, 106699: 0.0526702, 106700: 0.0649955, 106701: 0.125886, 106702: 0.0967257, 106703: 0.0706206, 106704: 0.0799989, 106705: 0.0847513, 106706: 0.135571, 106707: 0.0649855, 106708: 0.0492589, 106709: 0.0649172, 106710: 0.121263, 106711: 0.0670809, 106712: 0.0608073, 106713: 0.116288, 106714: 0.071609},
'Nuclei in individual cell region Selected - Nucleus HOECHST 33342 Haralick Homogeneity 1 px': {106695: 0.657532, 106696: 0.546708, 106697: 0.72884, 106698: 0.721774, 106699: 0.700476, 106700: 0.696009, 106701: 0.618728, 106702: 0.59469, 106703: 0.693487, 106704: 0.647874, 106705: 0.678351, 106706: 0.528893, 106707: 0.706147, 106708: 0.72233, 106709: 0.714676, 106710: 0.605918, 106711: 0.700766, 106712: 0.691383, 106713: 0.646318, 106714: 0.70725},
'Nuclei in individual cell region Selected - Nucleus HOECHST 33342 SER Spot 0 px': {106695: 0.00861118, 106696: 0.00931817, 106697: 0.00761309, 106698: 0.00837558, 106699: 0.0082335, 106700: 0.00795943, 106701: 0.00823312, 106702: 0.00783509, 106703: 0.00730663, 106704: 0.00735734, 106705: 0.00698037, 106706: 0.00857095, 106707: 0.007307, 106708: 0.00651859, 106709: 0.00674888, 106710: 0.00777671, 106711: 0.00729998, 106712: 0.00619496, 106713: 0.00603798, 106714: 0.0066989},
'Nuclei in individual cell region Selected - Nucleus HOECHST 33342 SER Hole 0 px': {106695: 0.00781875, 106696: 0.00746205, 106697: 0.00702571, 106698: 0.00721342, 106699: 0.00711043, 106700: 0.00706697, 106701: 0.00467969, 106702: 0.00478292, 106703: 0.00639713, 106704: 0.00709484, 106705: 0.00655664, 106706: 0.00715089, 106707: 0.00645719, 106708: 0.00597439, 106709: 0.00616917, 106710: 0.00496998, 106711: 0.00638658, 106712: 0.00532789, 106713: 0.00529905, 106714: 0.00612883},
'Nuclei in individual cell region Selected - Nucleus HOECHST 33342 SER Edge 0 px': {106695: 0.0729417, 106696: 0.110059, 106697: 0.0569585, 106698: 0.0598186, 106699: 0.0642045, 106700: 0.0669293, 106701: 0.0972561, 106702: 0.0924378, 106703: 0.0656912, 106704: 0.0757634, 106705: 0.069061, 106706: 0.111463, 106707: 0.063571, 106708: 0.0612379, 106709: 0.0594756, 106710: 0.09432, 106711: 0.065867, 106712: 0.0676253, 106713: 0.0765422, 106714: 0.0634227},
'Nuclei in individual cell region Selected - Nucleus HOECHST 33342 SER Ridge 0 px': {106695: 0.0149807, 106696: 0.0148023, 106697: 0.0134511, 106698: 0.013989, 106699: 0.0136802, 106700: 0.0135172, 106701: 0.0128792, 106702: 0.0118276, 106703: 0.0124749, 106704: 0.0131911, 106705: 0.0119413, 106706: 0.0147721, 106707: 0.012416, 106708: 0.0114262, 106709: 0.0113361, 106710: 0.0129007, 106711: 0.0124422, 106712: 0.010958, 106713: 0.0110026, 106714: 0.0118087},
'Nuclei in individual cell region Selected - Nucleus HOECHST 33342 SER Valley 0 px': {106695: 0.0161717, 106696: 0.0220035, 106697: 0.0138008, 106698: 0.0147323, 106699: 0.0145775, 106700: 0.0143745, 106701: 0.0137458, 106702: 0.0146674, 106703: 0.0141544, 106704: 0.0154375, 106705: 0.013253, 106706: 0.0246233, 106707: 0.0128277, 106708: 0.012231, 106709: 0.0126041, 106710: 0.013144, 106711: 0.0138948, 106712: 0.0126162, 106713: 0.0149189, 106714: 0.0139237},
'Nuclei in individual cell region Selected - Nucleus HOECHST 33342 SER Saddle 0 px': {106695: 0.0164057, 106696: 0.0177186, 106697: 0.0141956, 106698: 0.0141493, 106699: 0.0148899, 106700: 0.0142285, 106701: 0.0138838, 106702: 0.0152582, 106703: 0.013782, 106704: 0.0151764, 106705: 0.0132659, 106706: 0.0180964, 106707: 0.013406, 106708: 0.0118983, 106709: 0.0130469, 106710: 0.0133371, 106711: 0.0139153, 106712: 0.0121744, 106713: 0.0139629, 106714: 0.013405},
'Nuclei in individual cell region Selected - Nucleus HOECHST 33342 SER Bright 0 px': {106695: 0.0205846, 106696: 0.0209992, 106697: 0.0183712, 106698: 0.019501, 106699: 0.0190916, 106700: 0.0187158, 106701: 0.0183655, 106702: 0.0170604, 106703: 0.01726, 106704: 0.0179373, 106705: 0.0164909, 106706: 0.0203458, 106707: 0.0171765, 106708: 0.0156539, 106709: 0.0157577, 106710: 0.0180485, 106711: 0.0172014, 106712: 0.0149629, 106713: 0.0148986, 106714: 0.0161332},
'Nuclei in individual cell region Selected - Nucleus HOECHST 33342 SER Dark 0 px': {106695: 0.0209977, 106696: 0.0260261, 106697: 0.0182116, 106698: 0.0192058, 106699: 0.0189864, 106700: 0.0187439, 106701: 0.0162118, 106702: 0.017106, 106703: 0.0180129, 106704: 0.0197488, 106705: 0.0173244, 106706: 0.0281423, 106707: 0.0168369, 106708: 0.0159467, 106709: 0.0164385, 106710: 0.0159221, 106711: 0.0177785, 106712: 0.0157466, 106713: 0.0177876, 106714: 0.0176109},
'Nuclei in individual cell region Selected - Intensity Nucleus HOECHST 33342 Mean': {106695: 11255.9, 106696: 26866.3, 106697: 10728.4, 106698: 9493.33, 106699: 11029.7, 106700: 10894.4, 106701: 40807.4, 106702: 33047.2, 106703: 11523.0, 106704: 14381.5, 106705: 11346.3, 106706: 26054.4, 106707: 12701.5, 106708: 9906.32, 106709: 14341.3, 106710: 26745.4, 106711: 11314.5, 106712: 12563.2, 106713: 13698.5, 106714: 11522.0},
'Nuclei in individual cell region Selected - Intensity Nucleus HOECHST 33342 StdDev': {106695: 3365.55, 106696: 10684.8, 106697: 2379.63, 106698: 2439.61, 106699: 2738.63, 106700: 2988.91, 106701: 15826.6, 106702: 11332.9, 106703: 3275.83, 106704: 4358.66, 106705: 3512.88, 106706: 10206.0, 106707: 3486.11, 106708: 2382.69, 106709: 3903.72, 106710: 10256.8, 106711: 3153.22, 106712: 3386.94, 106713: 4945.58, 106714: 3311.51},
'Nuclei in individual cell region Selected - Individual Cell Region resized Area [µm²]': {106695: 473.462, 106696: 774.458, 106697: 1080.01, 106698: 775.12, 106699: 734.379, 106700: 491.614, 106701: 129.6, 106702: 211.134, 106703: 549.947, 106704: 359.89, 106705: 548.911, 106706: 564.429, 106707: 409.792, 106708: 637.515, 106709: 525.013, 106710: 146.614, 106711: 479.139, 106712: 357.323, 106713: 302.253, 106714: 377.957},
'Nuclei in individual cell region Selected - Individual Cell Region resized Roundness': {106695: 0.808892, 106696: 0.804004, 106697: 0.913968, 106698: 0.86118, 106699: 0.89257, 106700: 0.882859, 106701: 0.559616, 106702: 0.84152, 106703: 0.860951, 106704: 0.939933, 106705: 0.912447, 106706: 0.778607, 106707: 0.892991, 106708: 0.855366, 106709: 0.729886, 106710: 0.782424, 106711: 0.901534, 106712: 0.907393, 106713: 0.855864, 106714: 0.821204},
'Nuclei in individual cell region Selected - Individual Cell Region resized Width [µm]': {106695: 17.961, 106696: 24.8965, 106697: 30.1663, 106698: 25.6286, 106699: 23.994, 106700: 20.1043, 106701: 6.52379, 106702: 13.5987, 106703: 21.1186, 106704: 16.9182, 106705: 22.8623, 106706: 21.0169, 106707: 19.2897, 106708: 23.6516, 106709: 16.6386, 106710: 10.5238, 106711: 21.0928, 106712: 19.1464, 106713: 15.8732, 106714: 13.3183},
'Nuclei in individual cell region Selected - Individual Cell Region resized Length [µm]': {106695: 33.1711, 106696: 38.3933, 106697: 43.0947, 106698: 39.4182, 106699: 37.0375, 106700: 31.0506, 106701: 20.972, 106702: 20.0319, 106703: 33.7673, 106704: 28.7935, 106705: 28.5463, 106706: 36.9713, 106707: 29.5297, 106708: 32.7235, 106709: 38.1401, 106710: 16.6881, 106711: 30.8736, 106712: 24.0524, 106713: 24.5909, 106714: 32.0091},
'Nuclei in individual cell region Selected - Individual Cell Region resized Ratio Width to Length': {106695: 0.541467, 106696: 0.648461, 106697: 0.700001, 106698: 0.650171, 106699: 0.647832, 106700: 0.647469, 106701: 0.311071, 106702: 0.678853, 106703: 0.625416, 106704: 0.587569, 106705: 0.800884, 106706: 0.568466, 106707: 0.653231, 106708: 0.72277, 106709: 0.43625, 106710: 0.630619, 106711: 0.6832, 106712: 0.79603, 106713: 0.645494, 106714: 0.41608},
'Nuclei in individual cell region Selected - Relative Spot Intensity': {106695: 0.053115, 106696: 0.030453, 106697: 0.0528771, 106698: 0.0706828, 106699: 0.0553709, 106700: 0.0548624, 106701: 0.0991606, 106702: 0.0846535, 106703: 0.0676428, 106704: 0.138471, 106705: 0.0741397, 106706: 0.0459002, 106707: 0.0422811, 106708: 0.0763994, 106709: 0.0122011, 106710: 0.020017, 106711: 0.0777289, 106712: 0.0340526, 106713: 0.0368442, 106714: 0.0485223},
'Nuclei in individual cell region Selected - Number of Spots per Area of Individual Cell Region resized': {106695: 0.00107697, 106696: 0.00052672, 106697: 0.000865569, 106698: 0.0009429, 106699: 0.000833198, 106700: 0.000898907, 106701: 0.00170492, 106702: 0.000885526, 106703: 0.00108172, 106704: 0.00207802, 106705: 0.00105279, 106706: 0.000451698, 106707: 0.000829531, 106708: 0.000906473, 106709: 0.000258992, 106710: 0.000231857, 106711: 0.00106421, 106712: 0.000570803, 106713: 0.000843502, 106714: 0.000629581},
'Compound': {106695: 'Ctrl', 106696: 'Ctrl', 106697: 'Ctrl', 106698: 'Ctrl', 106699: 'Ctrl', 106700: 'Ctrl', 106701: 'Ctrl', 106702: 'Ctrl', 106703: 'Ctrl', 106704: 'Ctrl', 106705: 'Ctrl', 106706: 'Ctrl', 106707: 'Ctrl', 106708: 'Ctrl', 106709: 'Ctrl', 106710: 'Ctrl', 106711: 'Ctrl', 106712: 'Ctrl', 106713: 'Ctrl', 106714: 'Ctrl'}}
df1_Control = pd.DataFrame(df_Control)
I have a data set for many drugs (Compounds) with many features (columns). I want to make a loop to generate histograms from each column of df_Sample for each drug, side by side in a facetgrid. In addition, for comparison, I need to take the data from same columns in the df_Control and merge it on the appropriate histogram of df_Sample. When I just want to have df_Sample histograms, I get the desired results from the code below:
i = 0
for i, column, in enumerate(df1_Sample.columns):
sns.FacetGrid(data=df1_Control, col='Compound', col_wrap= 6).map(plt.hist, column)
file_name = 'plot_' column '.png'
plt.savefig(file_name)
However, cannot make the script work for merging the samples with the relevant control on the same graph. I thought maybe someone can revise the script that came to my mind:
i1 = 0
i2 = 0
for (i1, column1), (i2, column2) in zip(enumerate(df1_Sample.columns), enumerate(df1_Sample.columns)):
sns.FacetGrid(data=[df1_Sample, df1_Control], col='Compound', col_wrap= 6).map(plt.hist, column) #In FaceGrid, use col for determining the identifier, which is the name of the compounds.
sns.FacetGrid(data=df1_Control, col='Compound', col_wrap= 6).map(plt.hist, column)
plt.xlabel("Data", size=14)
plt.ylabel("Count", size=14)
plt.legend(loc='upper right')
file_name = 'plot_' column '.png'
plt.savefig(file_name, dpi=1200)
Nevertheless, I don't know if can, for example, use 'fig, ax = plt.subplots()' somehow for this purpose or can make the 'sns.FacetGrid' work.
I very much appreciate your kind suggestion.
CodePudding user response:
-
- Response to comment about
ValueError: arrays must all be same length- Here is an alternate way to combine the data from the two dataframes, when the number of rows between the two dataframes is not the same.
- The issue becomes that the
'Compound'column indf1_Controlonly contains'Ctrl', so there's no way to associate the rows with the'Compound'fromdf1_Sample. So, all of the control data would be plotted in a separate histogram. - Therefore, the data in the
'Compound'column ofdf1_Control, needs to be properly labeled.
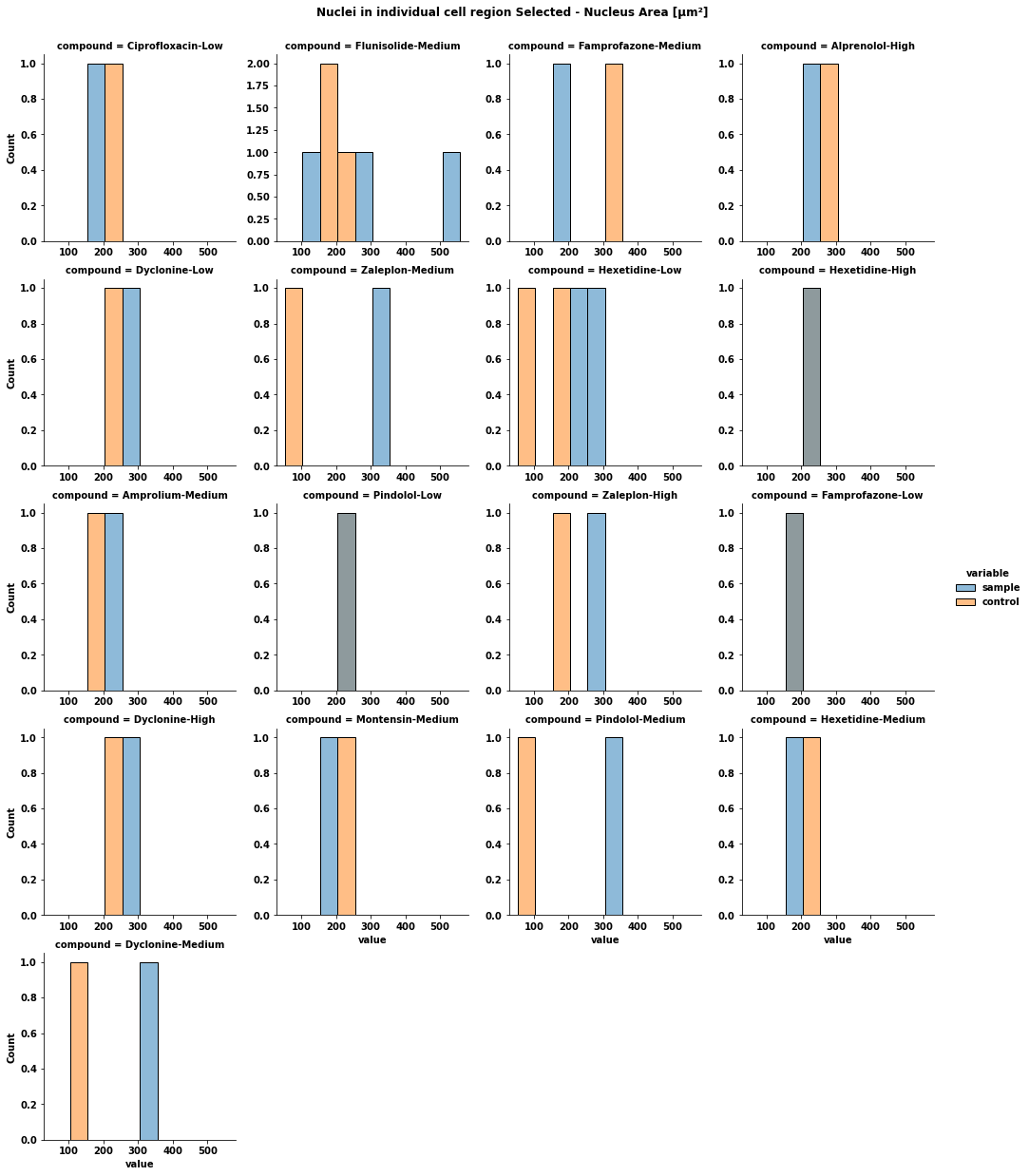
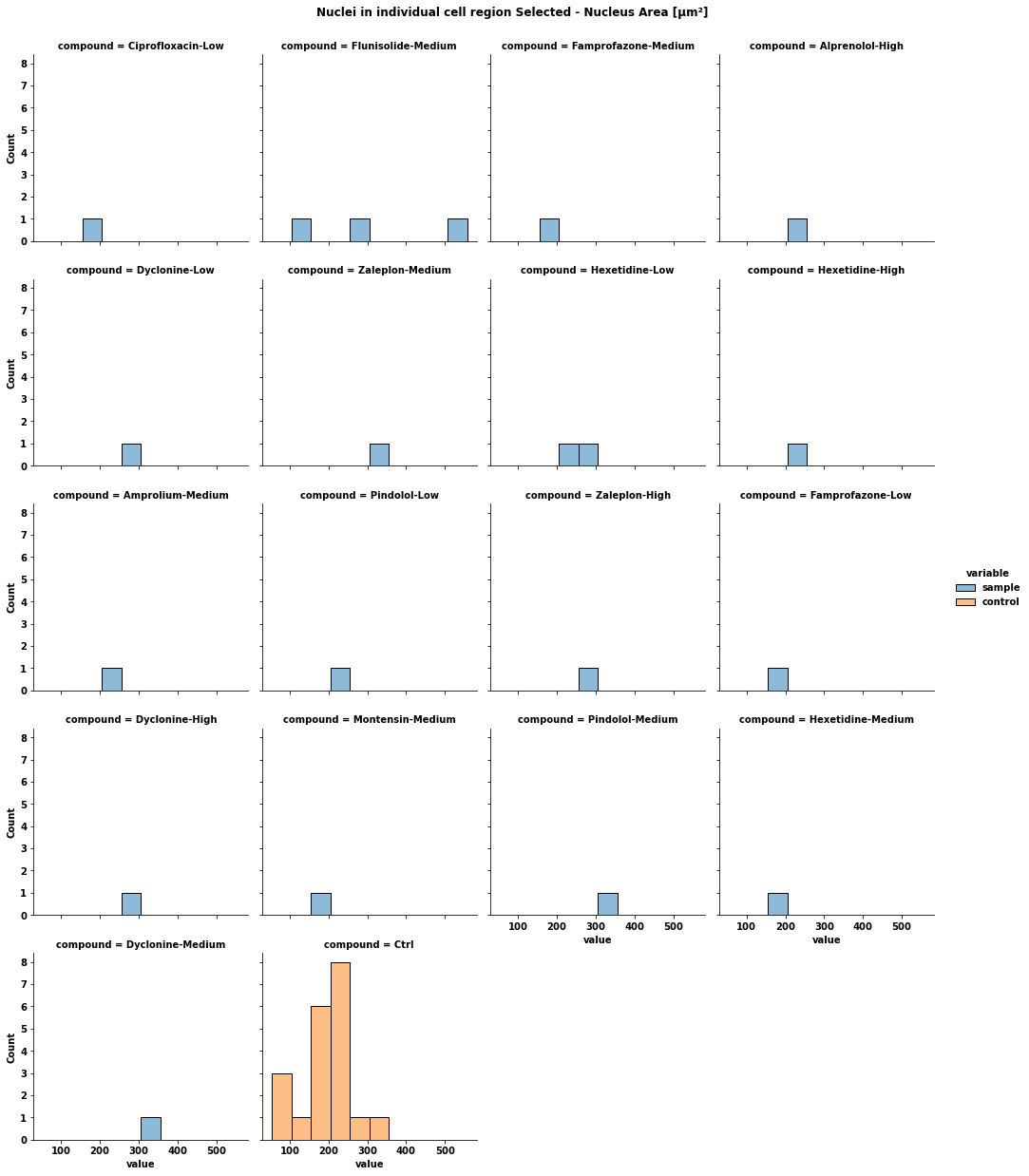
for col in df1_Sample.columns[:1]: # combine the data from sample and control sample = df1_Sample[[col, 'Compound']].copy() sample['variable'] = 'sample' control = df1_Control[[col, 'Compound']].copy() control['variable'] = 'control' data = pd.concat([sample, control]).reset_index(drop=True) data.columns = ['value', 'compound', 'variable'] display(data) # plot the data p = sns.displot(data=data, x='value', hue='variable', col='compound', col_wrap=4, height=3.5) p.fig.subplots_adjust(top=0.94) # adjust the Figure in p p.fig.suptitle(col) file_name = f'plot {col}.png' p.savefig(file_name) [out]: value compound variable 0 189.4800 Ciprofloxacin-Low sample 1 153.7360 Flunisolide-Medium sample 2 199.2190 Famprofazone-Medium sample 3 221.4000 Alprenolol-High sample 4 261.6480 Dyclonine-Low sample 5 304.0890 Flunisolide-Medium sample 6 345.9350 Zaleplon-Medium sample 7 218.9350 Hexetidine-Low sample 8 232.6010 Hexetidine-High sample 9 240.9120 Amprolium-Medium sample 10 208.1250 Pindolol-Low sample 11 260.7130 Zaleplon-High sample 12 161.1120 Famprofazone-Low sample 13 270.1810 Dyclonine-High sample 14 165.8880 Montensin-Medium sample 15 342.0770 Pindolol-Medium sample 16 158.3760 Hexetidine-Medium sample 17 557.0350 Flunisolide-Medium sample 18 319.9130 Dyclonine-Medium sample 19 257.2970 Hexetidine-Low sample 20 205.1850 Ctrl control 21 160.0080 Ctrl control 22 329.2270 Ctrl control 23 264.5210 Ctrl control 24 242.8670 Ctrl control 25 225.5980 Ctrl control 26 53.7438 Ctrl control 27 63.8908 Ctrl control 28 208.2440 Ctrl control 29 195.4800 Ctrl control 30 218.5100 Ctrl control 31 160.2620 Ctrl control 32 190.5680 Ctrl control 33 254.6970 Ctrl control 34 239.3990 Ctrl control 35 59.5907 Ctrl control 36 228.2670 Ctrl control 37 164.5120 Ctrl control 38 125.6910 Ctrl control 39 177.4120 Ctrl control- Since the control data is not labeled, for each compound, crate a control dataframe where all the data is tagged with the given compound. This would allow for comparing each compound against the distribution of all the control data for each column.
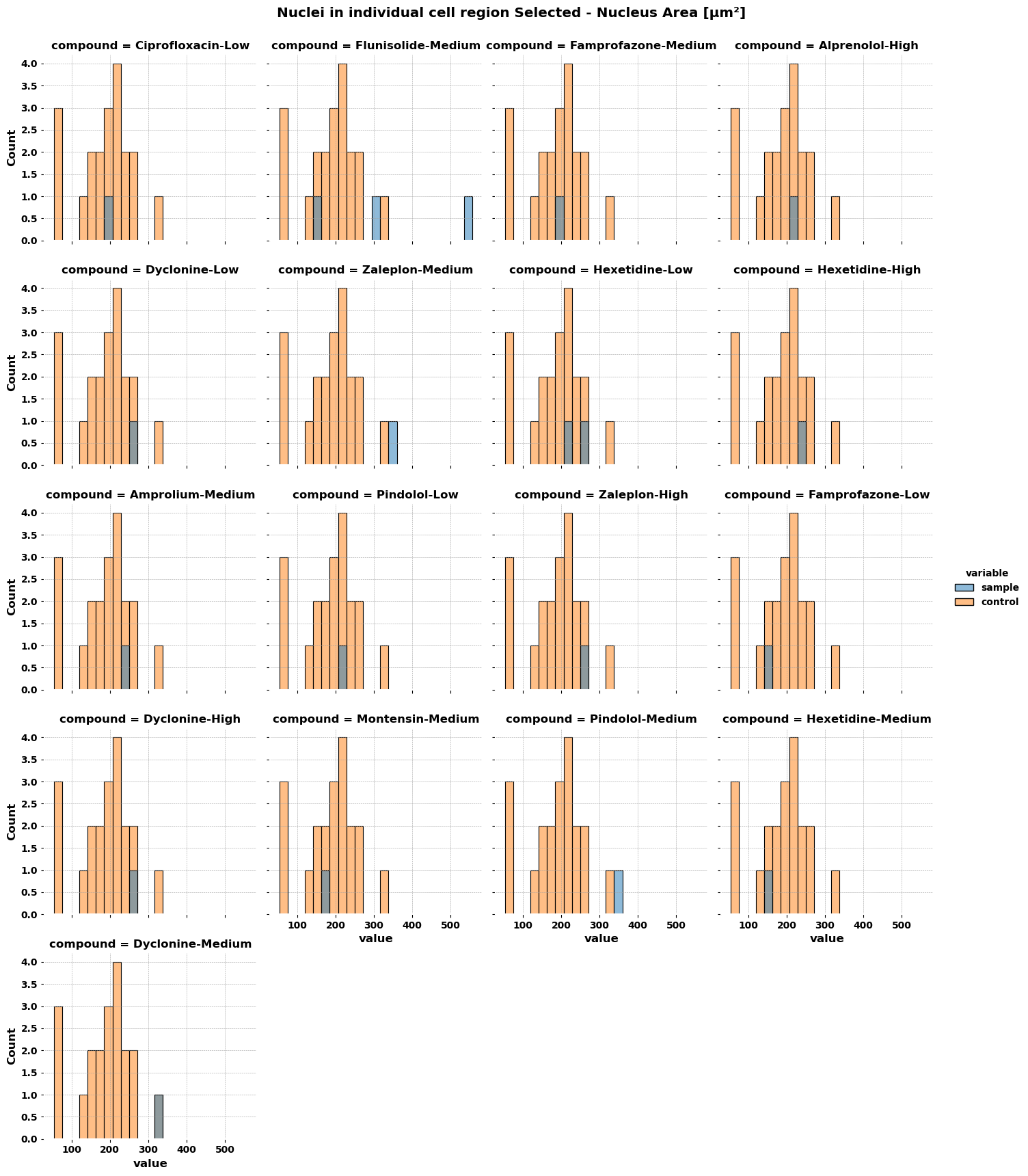
for col in df1_Sample.columns[:1]: # testing on first column; change to [:-1] for all but the last column # combine the data from sample and control sample = df1_Sample[[col, 'Compound']].copy() sample['variable'] = 'sample' control = df1_Control[[col]].copy() control['variable'] = 'control' compounds = df1_Sample['Compound'].unique() # for each compound, crate a control dataframe where all the data is tagged with the given compound control_list = list() for compound in compounds: ctrl = control.copy() ctrl['Compound'] = compound control_list.append(ctrl) data = pd.concat([sample] control_list).reset_index(drop=True) data.columns = ['value', 'compound', 'variable'] display(data.head()) # display works in a notebook, otherwise use print display(data.tail()) # remove or comment these display lines out # plot the data p = sns.displot(data=data, x='value', hue='variable', col='compound', col_wrap=4, height=3.5) p.fig.subplots_adjust(top=0.94) # adjust the Figure in p p.fig.suptitle(col) file_name = f'plot {col}.png' p.savefig(file_name) - Response to comment about