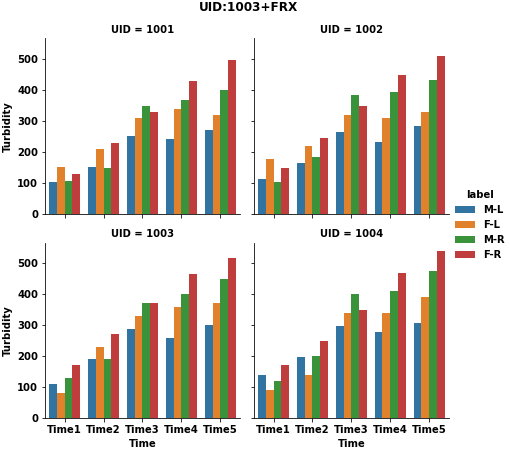
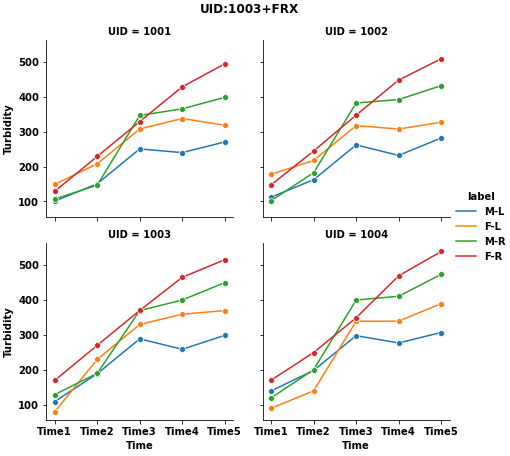
I have a dataframe df with 4 unique UID - 1001,1002,1003,1004.
I want to write a user-defined function in python that does the following:
- growth curve -plots
TurbidityagainstTimefor each uniqueUID.Turbidityvalues are the ones in theTime_1,Time_2,Time_3,Time_4&Time_5columns. For example,UID = 1003will have 4 plots on each graph
Add a legend to each graph such as
M L,F L,M R, andF R(from columnsGenandType)Add a title to each graph. For example-
UID:1003 Site:FRXExport the graphs as a
pdforjpegortifffile - 4 graphs per page
# The dataset
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
import matplotlib.patches as mpatches
import numpy as np
df= {
'Gen':['M','M','M','M','F','F','F','F','M','M','M','M','F','F','F','F'],
'Site':['FRX','FRX','FRX','FRX','FRX','FRX','FRX','FRX','FRX','FRX','FRX','FRX','FRX','FRX','FRX','FRX'],
'Type':['L','L','L','L','L','L','L','L','R','R','R','R','R','R','R','R'],
'UID':[1001,1002,1003,1004,1001,1002,1003,1004,1001,1002,1003,1004,1001,1002,1003,1004],
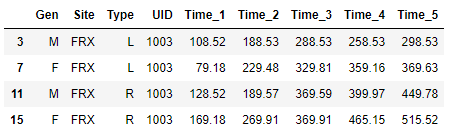
'Time1':[100.78,112.34,108.52,139.19,149.02,177.77,79.18,89.10,106.78,102.34,128.52,119.19,129.02,147.77,169.18,170.11],
'Time2':[150.78,162.34,188.53,197.69,208.07,217.76,229.48,139.51,146.87,182.54,189.57,199.97,229.28,244.73,269.91,249.19],
'Time3':[250.78,262.34,288.53,297.69,308.07,317.7,329.81,339.15,346.87,382.54,369.59,399.97,329.28,347.73,369.91,349.12],
'Time4':[240.18,232.14,258.53,276.69,338.07,307.74,359.16,339.25,365.87,392.48,399.97,410.75,429.08,448.39,465.15,469.33],
'Time5':[270.84,282.14,298.53,306.69,318.73,327.47,369.63,389.59,398.75,432.18,449.78,473.55,494.85,509.39,515.52,539.23]
}
df = pd.DataFrame(df,columns = ['Gen','Site','Type','UID','Time1','Time2','Time3','Time4','Time5'])
df
My attempt
# See below for my thoughts/attempt- I am open to other python libraries and approaches
def graph2pdf(inputdata):
#1. convert from wide to long
inputdata = pd.melt(df,id_vars = ['Gen','Type','UID'],var_name = 'Time',value_name = 'Turbidity')
#
cmaps = ['Reds', 'Blues', 'Greens', 'Greys','Yellows']
label_patches = []
for i, cmap in enumerate(cmaps):
# I want a growth curve not a distribution curve
sns.kdeplot(x = Time, y = Turbidity,data = data, cmap=cmaps[i] '_d')
label_patch = mpatches.Patch(color=sns.color_palette(cmaps[i])[2],label=label)
label_patches.append(label_patch)
#2. add legend
plt.legend(handles=label_patches, loc='upper left')
#3. add title- 'UID number SiteName: FRX' to each of the graphs
plt.title('UID:1003 FRX')
plt.show()
#4. export as pdf file i.e 4 graphs per page
with PdfPages('turbidityvstime_pdf.pdf') as pdf:
plt.figure(figsize=(2,2)) # 4 graphs per page, I am anticipating more pages in the future
pdf.savefig() # saves the current figure into a pdf page
plt.close()
# testing the user-defined function
graph2pdf(df)
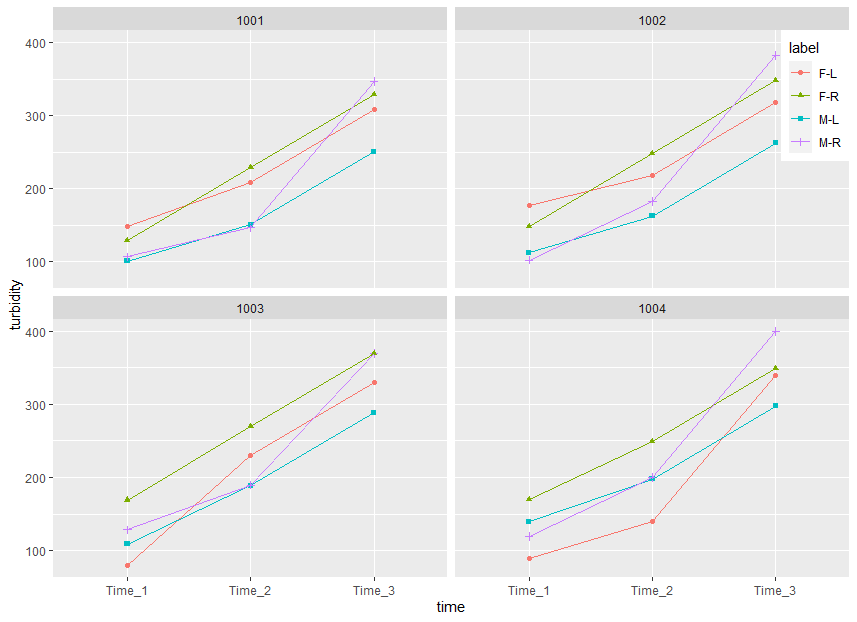
I want the graph to look something like the figure below (turbidity instead of density on the y-axis and time on the x-axis). if possible, a white or clear background is preferred
Thanks
CodePudding user response:
- I line plot is usually not appropriate for discrete data, because the slope of the lines can imply trends that do not exist.
- This is discrete because measurements are taken at discrete moments in time, not a continuous time series.
- Discrete data is best visualized with a bar plot.
- Use
seabornfigure-level methods like