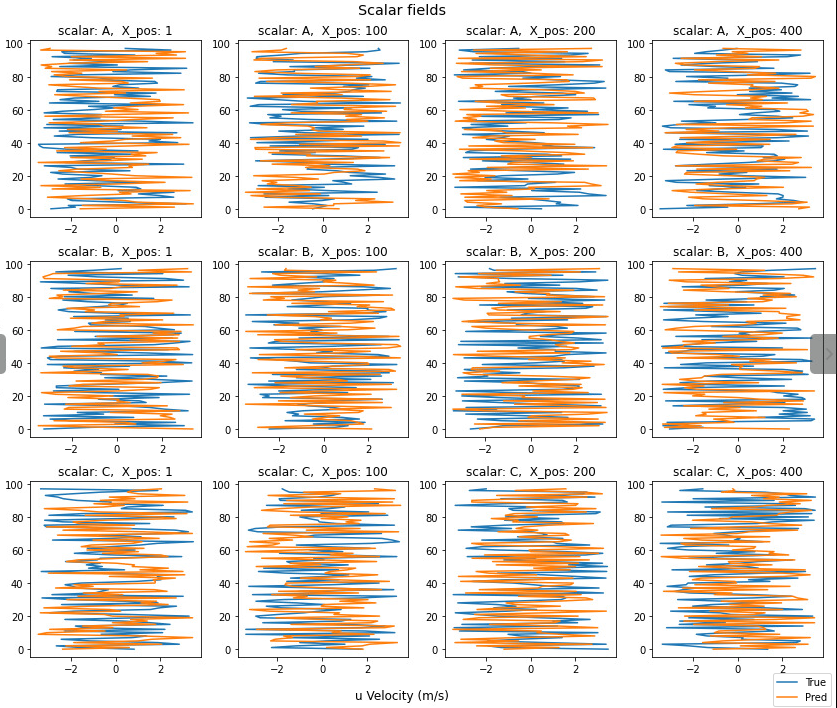
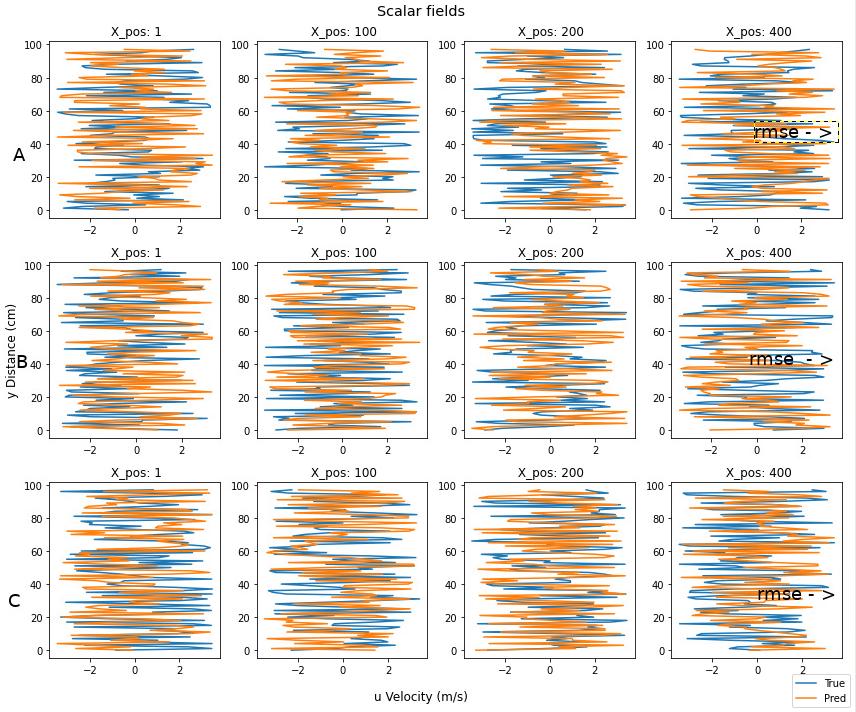
I've made the following function which creates scalar profile plots from a scalar field.
import numpy as np
import matplotlib.pyplot as plt
from sklearn.metrics import mean_squared_error
dummy_data_A = {'A': np.random.uniform(low=-3.5, high=3.5, size=(98,501)),
'B': np.random.uniform(low=-3.5, high=3.5, size=(98,501)), 'C': np.random.uniform(low=-3.5, high=3.5, size=(98,501))}
dummy_data_B = {'A': np.random.uniform(low=-3.5, high=3.5, size=(98,501)),
'B': np.random.uniform(low=-3.5, high=3.5, size=(98,501)), 'C': np.random.uniform(low=-3.5, high=3.5, size=(98,501))}
def plot_scalar_profiles(true_var, pred_var,
coordinates = ['x','y'],
x_position = None,
bounds_var = None,
norm = False ):
fig = plt.figure(figsize=(12,10))
st = plt.suptitle("Scalar fields", fontsize="x-large")
nr_plots = len(list(true_var.keys())) # not plotting x or y
plot_index = 1
for key in true_var.keys():
rmse_list = []
for profile_pos in x_position:
true_field_star = true_var[key]
pred_field_star = pred_var[key]
axes = plt.subplot(nr_plots, len(x_position), plot_index)
axes.set_title(f"scalar: {key}, X_pos: {profile_pos}") # ,RMSE: {rms:.2f}
true_profile = true_field_star[...,profile_pos] # for i in range(probe positions)
pred_profile = pred_field_star[...,profile_pos]
if norm:
#true_profile = true_field_star[...,profile_pos] # for i in range(probe positions)
#pred_profile = pred_field_star[...,profile_pos]
true_profile = normalize_list(true_field_star[...,profile_pos])
pred_profile = normalize_list(pred_field_star[...,profile_pos])
rms = mean_squared_error(true_profile, pred_profile, squared=False)
true_profile_y = range(len(true_profile))
axes.plot(true_profile, true_profile_y, label='True')
#
pred_profile_y = range(len(pred_profile))
axes.plot(pred_profile, pred_profile_y, label='Pred')
rmse_list.append(rms)
plot_index = 1
RMSE = sum(rmse_list)
print(f"RMSE {key}: {RMSE}")
lines, labels = fig.axes[-1].get_legend_handles_labels()
fig.legend(lines, labels, loc = "lower right")
fig.supxlabel('u Velocity (m/s)')
fig.supylabel('y Distance (cm)')
fig.tight_layout()
#plt.savefig(save_name '.png', facecolor='white', transparent=False)
plt.show()
plt.close('all')
plot_scalar_profiles(dummy_data_A, dummy_data_B,
x_position = [1, 100, 200, 400],
bounds_var=None,
norm=False)
It also computes the total root mean squared error of the profiles extracted along x_position for scalar fields 'A','B', 'C'.
Console output:
RMSE A: 11.815624240063709
RMSE B: 11.623509385371737
RMSE C: 11.435156749416366
Theres 2 things I have questions about:
How do I label each row and column? Labelling each subplot looks cluttered.
How to I annotate the RMSE outputs to the right of each row?
Here's a rough drawing of what I mean with the 2 points.
Also any other suggestions for improvements are welcome. I'm still trying to figure out whats the cleanest/best way to represent this data.
CodePudding user response:
Assumption
I base my answer on the hypothesis you have always 12 plots, so I focus my attention to some plots with fixed positions.
Answer
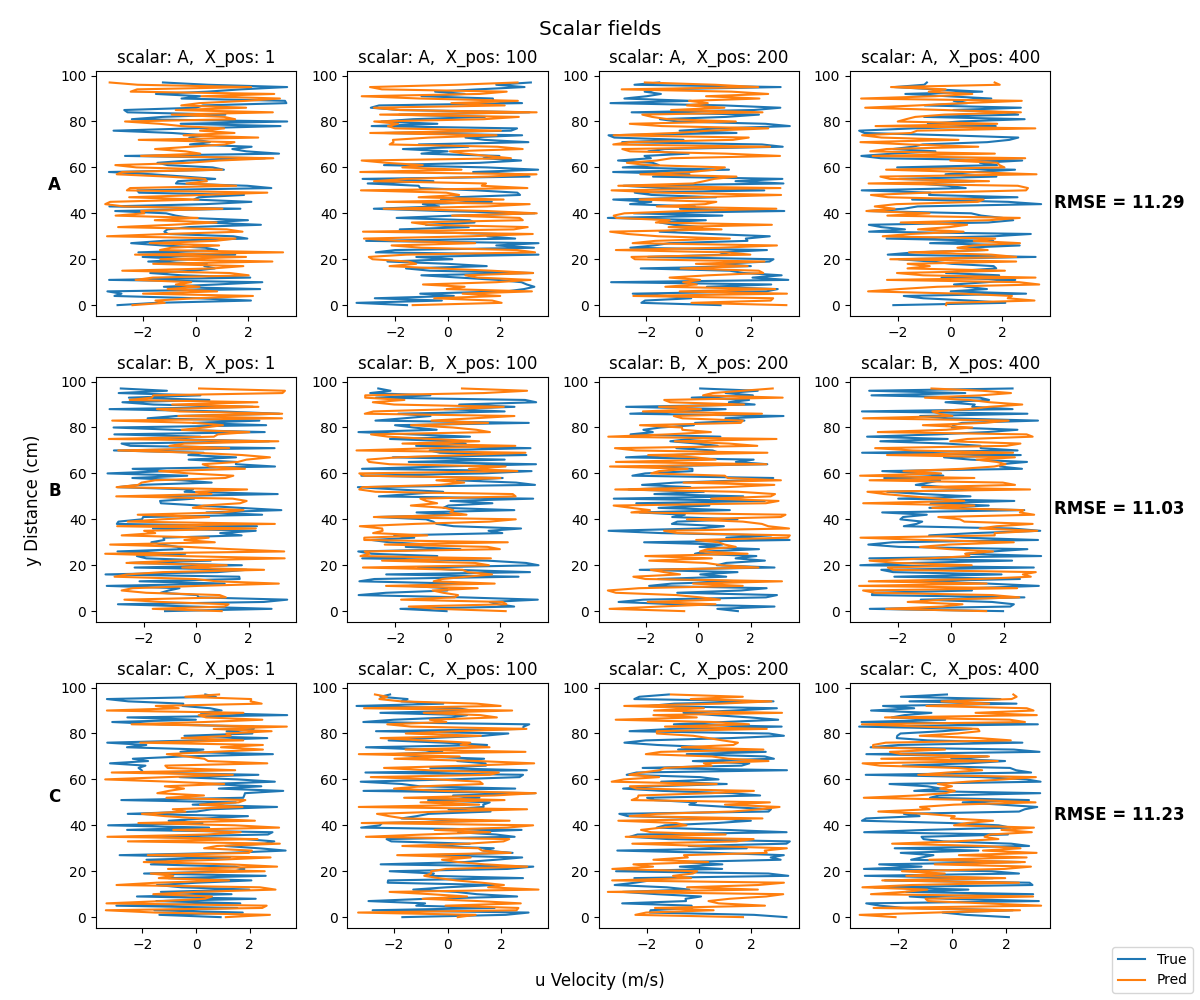
In order to label each row, you can set the y label of the plots on the left side with fig.axes[n].set_ylabel, optionally you can add additional parameter to customize the label:
fig.axes[0].set_ylabel('A', rotation = 0, weight = 'bold', fontsize = 12)
fig.axes[4].set_ylabel('B', rotation = 0, weight = 'bold', fontsize = 12)
fig.axes[8].set_ylabel('C', rotation = 0, weight = 'bold', fontsize = 12)
In order to report RMSE for each row, you need to save those values in a list, then you can exploit the same concept as above: set the y label of the plots on the right side and move the y label to the right:
RMSE = sum(rmse_list)
RMSE_values.append(RMSE)
print(f"RMSE {key}: {RMSE}")
...
fig.axes[3].set_ylabel(f'RMSE = {RMSE_values[0]:.2f}', rotation = 0, weight = 'bold', fontsize = 12, labelpad = 50)
fig.axes[7].set_ylabel(f'RMSE = {RMSE_values[1]:.2f}', rotation = 0, weight = 'bold', fontsize = 12, labelpad = 50)
fig.axes[11].set_ylabel(f'RMSE = {RMSE_values[2]:.2f}', rotation = 0, weight = 'bold', fontsize = 12, labelpad = 50)
fig.axes[3].yaxis.set_label_position('right')
fig.axes[7].yaxis.set_label_position('right')
fig.axes[11].yaxis.set_label_position('right')
Complete Code
import numpy as np
import matplotlib.pyplot as plt
from sklearn.metrics import mean_squared_error
dummy_data_A = {'A': np.random.uniform(low = -3.5, high = 3.5, size = (98, 501)),
'B': np.random.uniform(low = -3.5, high = 3.5, size = (98, 501)),
'C': np.random.uniform(low = -3.5, high = 3.5, size = (98, 501))}
dummy_data_B = {'A': np.random.uniform(low = -3.5, high = 3.5, size = (98, 501)),
'B': np.random.uniform(low = -3.5, high = 3.5, size = (98, 501)),
'C': np.random.uniform(low = -3.5, high = 3.5, size = (98, 501))}
def plot_scalar_profiles(true_var, pred_var,
coordinates = ['x', 'y'],
x_position = None,
bounds_var = None,
norm = False):
fig = plt.figure(figsize = (12, 10))
st = plt.suptitle("Scalar fields", fontsize = "x-large")
nr_plots = len(list(true_var.keys())) # not plotting x or y
plot_index = 1
RMSE_values = []
for key in true_var.keys():
rmse_list = []
for profile_pos in x_position:
true_field_star = true_var[key]
pred_field_star = pred_var[key]
axes = plt.subplot(nr_plots, len(x_position), plot_index)
axes.set_title(f"scalar: {key}, X_pos: {profile_pos}") # ,RMSE: {rms:.2f}
true_profile = true_field_star[..., profile_pos] # for i in range(probe positions)
pred_profile = pred_field_star[..., profile_pos]
if norm:
# true_profile = true_field_star[...,profile_pos] # for i in range(probe positions)
# pred_profile = pred_field_star[...,profile_pos]
true_profile = normalize_list(true_field_star[..., profile_pos])
pred_profile = normalize_list(pred_field_star[..., profile_pos])
rms = mean_squared_error(true_profile, pred_profile, squared = False)
true_profile_y = range(len(true_profile))
axes.plot(true_profile, true_profile_y, label = 'True')
#
pred_profile_y = range(len(pred_profile))
axes.plot(pred_profile, pred_profile_y, label = 'Pred')
rmse_list.append(rms)
plot_index = 1
RMSE = sum(rmse_list)
RMSE_values.append(RMSE)
print(f"RMSE {key}: {RMSE}")
lines, labels = fig.axes[-1].get_legend_handles_labels()
fig.legend(lines, labels, loc = "lower right")
fig.supxlabel('u Velocity (m/s)')
fig.supylabel('y Distance (cm)')
fig.axes[0].set_ylabel('A', rotation = 0, weight = 'bold', fontsize = 12)
fig.axes[4].set_ylabel('B', rotation = 0, weight = 'bold', fontsize = 12)
fig.axes[8].set_ylabel('C', rotation = 0, weight = 'bold', fontsize = 12)
fig.axes[3].set_ylabel(f'RMSE = {RMSE_values[0]:.2f}', rotation = 0, weight = 'bold', fontsize = 12, labelpad = 50)
fig.axes[7].set_ylabel(f'RMSE = {RMSE_values[1]:.2f}', rotation = 0, weight = 'bold', fontsize = 12, labelpad = 50)
fig.axes[11].set_ylabel(f'RMSE = {RMSE_values[2]:.2f}', rotation = 0, weight = 'bold', fontsize = 12, labelpad = 50)
fig.axes[3].yaxis.set_label_position('right')
fig.axes[7].yaxis.set_label_position('right')
fig.axes[11].yaxis.set_label_position('right')
fig.tight_layout()
# plt.savefig(save_name '.png', facecolor='white', transparent=False)
plt.show()
plt.close('all')
plot_scalar_profiles(dummy_data_A, dummy_data_B,
x_position = [1, 100, 200, 400],
bounds_var = None,
norm = False)