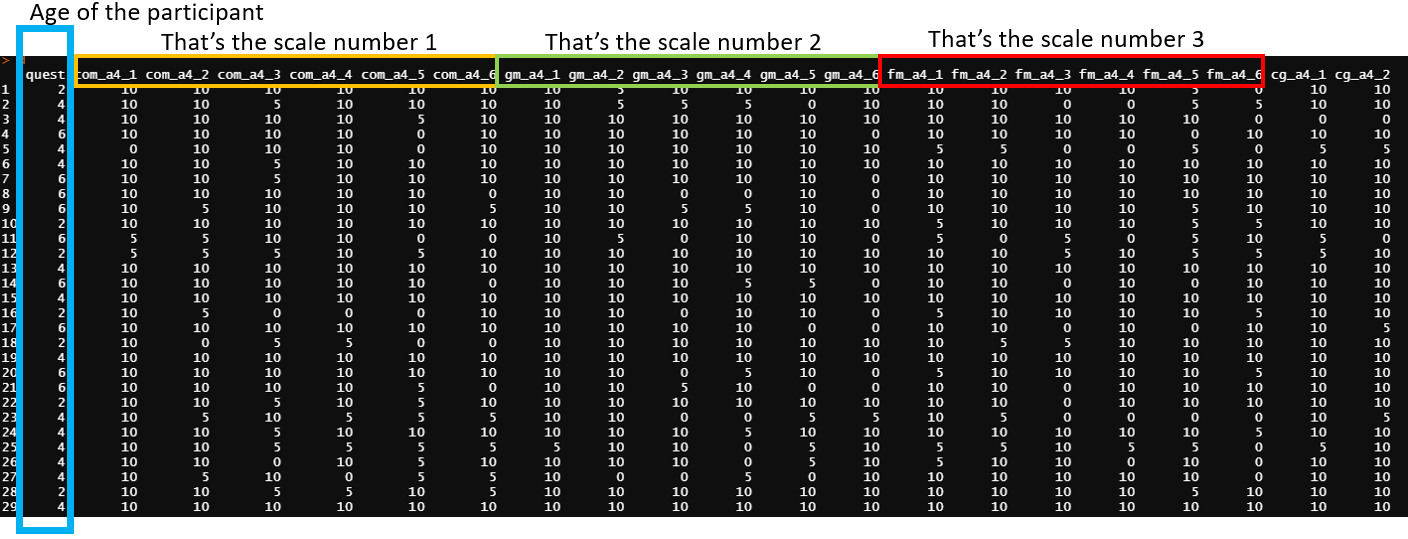
In psychology, this kind of dataset presented below is pretty common
I would like to group all age (variable = quest), than group all scales (com_a4_1:com_a4_6; and gm_a4_1:gm_a4_6, etc) and then apply a reliability function to the data (psych::alpha).
I successfully create this syntax
d %>%
select(quest,contains("_a4_")) %>% #get the data
group_by(quest) %>% #group by all age interval
do(alpha(.)$total)
However, I'm not being able to "sub" nest using the scales' items.
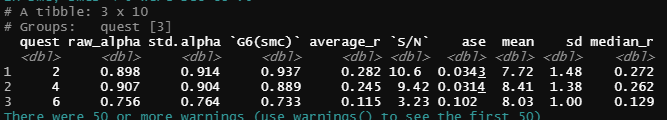
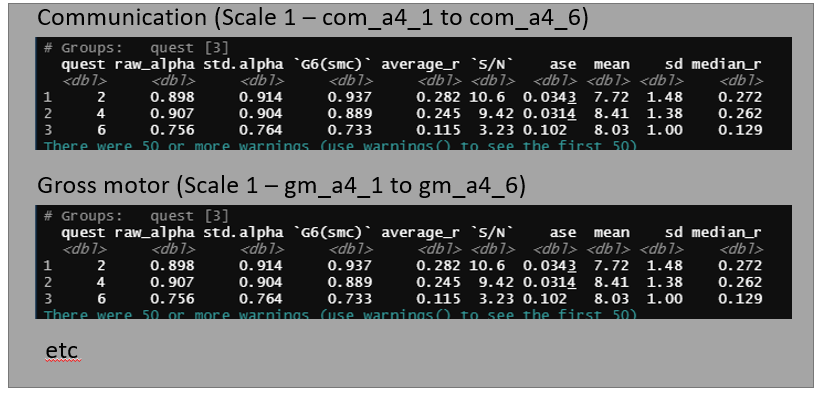
As far as I imagine, I'll have to pivot my data and then group or nest. However, I'm not having any success at this point. My expected result is something similar to this image below. There are "two nested results". The first result is grouped with the scale (ex: com_a4_1:com_a4_6) and the second is grouped with the age (quest)
Fake data and codes are below
library(psych)
library(tidyverse)
d %>%
select(quest,contains("_a4_")) %>% #get the data
group_by(quest) %>% #group by all age interval
do(alpha(.)$total)
d <-structure(list(quest = c(6, 4, 2, 4, 2, 6, 2, 4, 2, 2, 4, 2,
6, 4, 4, 2, 2, 4, 2, 6, 2, 2, 4, 6, 6, 4, 4, 4, 2, 6, 4, 2, 6,
4, 6, 2, 2, 4, 6, 4, 2), com_a4_1 = c(10, 0, 10, 10, 5, 10, 5,
10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 5, 10, 10, 0, 10,
10, 10, 10, 10, 5, 10, 10, 10, 10, 10, 10, 10, 10, 5, 10, 10,
10, 10), com_a4_2 = c(10, 10, 5, 10, 10, 5, 10, 10, 10, 10, 10,
10, 10, 10, 10, 10, 10, 10, 10, 10, 5, 5, 10, 10, 10, 10, 5,
10, 10, 10, 5, 0, 10, 10, 10, 10, 0, 10, 10, 10, 10), com_a4_3 = c(10,
5, 0, 5, 10, 5, 5, 10, 10, 10, 10, 10, 5, 5, 10, 10, 5, 10, 10,
10, 10, 5, 5, 10, 10, 5, 5, 10, 10, 10, 10, 5, 10, 10, 10, 10,
0, 10, 5, 10, 10), com_a4_4 = c(10, 0, 0, 10, 5, 10, 10, 10,
10, 5, 5, 10, 10, 5, 10, 10, 5, 10, 10, 10, 10, 5, 10, 10, 10,
10, 0, 10, 5, 10, 10, 10, 10, 10, 10, 10, 5, 10, 10, 10, 10),
com_a4_5 = c(10, 0, 0, 5, 0, 10, 5, 10, 10, 5, 10, 10, 0,
10, 10, 10, 0, 10, 5, 10, 0, 0, 10, 0, 10, 10, 10, 10, 5,
0, 10, 5, 5, 10, 10, 10, 0, 10, 10, 10, 10), com_a4_6 = c(5,
10, 0, 10, 10, 5, 10, 10, 10, 0, 10, 10, 5, 10, 10, 10, 10,
10, 10, 5, 10, 10, 10, 10, 10, 10, 10, 10, 5, 10, 5, 10,
5, 10, 5, 10, 0, 10, 5, 10, 10), gm_a4_1 = c(10, 10, 10,
10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10,
10, 10, 10, 10, 10, 10, 5, 10, 10, 10, 10, 10, 10, 10, 10,
10, 10, 10, 10, 10, 10, 10, 10), gm_a4_2 = c(10, 10, 10,
10, 10, 10, 10, 10, 10, 10, 10, 10, 5, 10, 10, 10, 10, 10,
10, 10, 10, 10, 10, 10, 10, 5, 5, 10, 10, 10, 0, 10, 10,
5, 10, 10, 5, 10, 10, 10, 10), gm_a4_3 = c(10, 10, 10, 10,
10, 5, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10,
10, 10, 10, 10, 10, 10, 0, 0, 10, 10, 10, 0, 10, 10, 10,
10, 10, 5, 10, 10, 10, 10), gm_a4_4 = c(0, 5, 10, 10, 10,
10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 5,
10, 10, 10, 10, 10, 0, 0, 10, 10, 10, 0, 10, 5, 5, 5, 10,
10, 10, 10, 10, 10), gm_a4_5 = c(10, 10, 10, 10, 10, 10,
10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10,
10, 10, 10, 10, 10, 10, 10, 10, 10, 5, 10, 10, 10, 5, 10,
5, 10, 10, 10, 10), gm_a4_6 = c(0, 10, 5, 5, 10, 5, 5, 10,
10, 5, 10, 10, 0, 10, 10, 10, 5, 10, 5, 10, 10, 10, 10, 0,
10, 10, 10, 10, 10, 0, 10, 10, 10, 10, 0, 10, 0, 10, 10,
10, 10), fm_a4_1 = c(10, 5, 10, 10, 10, 10, 10, 10, 10, 10,
10, 10, 10, 10, 10, 5, 10, 10, 10, 10, 5, 0, 10, 10, 0, 5,
10, 10, 10, 10, 5, 5, 10, 10, 5, 5, 10, 10, 10, 10, 10),
fm_a4_2 = c(10, 10, 10, 10, 0, 10, 10, 10, 10, 10, 10, 10,
10, 10, 10, 10, 10, 10, 10, 5, 10, 10, 10, 10, 10, 10, 5,
10, 10, 5, 10, 10, 10, 10, 5, 10, 10, 10, 10, 10, 10), fm_a4_3 = c(0,
5, 10, 10, 5, 10, 5, 10, 10, 10, 10, 10, 5, 10, 5, 5, 5,
10, 10, 5, 0, 10, 5, 10, 5, 10, 10, 0, 10, 10, 5, 10, 10,
10, 0, 10, 0, 10, 10, 10, 10), fm_a4_4 = c(10, 5, 10, 10,
10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10,
10, 10, 10, 5, 10, 10, 10, 5, 10, 10, 10, 0, 10, 10, 10,
10, 10, 0, 10, 10, 10, 10), fm_a4_5 = c(0, 5, 10, 10, 10,
0, 10, 10, 10, 10, 10, 10, 0, 10, 10, 5, 10, 10, 5, 0, 10,
10, 10, 10, 10, 10, 5, 10, 10, 0, 5, 10, 0, 10, 0, 5, 5,
5, 10, 10, 10), fm_a4_6 = c(10, 5, 5, 0, 0, 5, 10, 10, 10,
0, 10, 10, 5, 10, 10, 10, 0, 10, 0, 10, 10, 0, 10, 10, 5,
0, 0, 10, 10, 10, 0, 10, 10, 5, 5, 10, 0, 0, 10, 10, 5),
cg_a4_1 = c(10, 5, 10, 5, 10, 10, 10, 10, 10, 10, 10, 10,
10, 10, 10, 10, 10, 10, 10, 10, 0, 10, 10, 10, 10, 5, 0,
10, 10, 10, 10, 5, 10, 10, 10, 10, 5, 5, 10, 10, 10), cg_a4_2 = c(5,
10, 10, 5, 10, 5, 10, 10, 10, 10, 10, 10, 5, 10, 10, 10,
10, 10, 10, 5, 10, 10, 10, 10, 10, 5, 10, 10, 10, 10, 10,
10, 10, 10, 10, 10, 10, 10, 10, 10, 10), cg_a4_3 = c(10,
10, 5, 10, 10, 10, 10, 10, 10, 5, 10, 10, 5, 10, 10, 10,
5, 10, 10, 10, 10, 0, 10, 10, 5, 10, 5, 10, 10, 10, 5, 10,
10, 10, 10, 10, 5, 10, 10, 10, 10), cg_a4_4 = c(10, 10, 0,
5, 5, 5, 10, 10, 10, 5, 10, 10, 0, 5, 10, 10, 5, 10, 10,
10, 10, 0, 5, 10, 10, 5, 0, 0, 10, 10, 0, 10, 0, 10, 10,
5, 0, 5, 5, 10, 10), cg_a4_5 = c(5, 0, 0, 5, 0, 10, 5, 10,
10, 0, 10, 10, 10, 10, 5, 10, 0, 10, 0, 10, 0, 0, 10, 10,
5, 10, 5, 10, 5, 5, 5, 0, 10, 10, 5, 10, 0, 10, 10, 10, 10
), cg_a4_6 = c(0, 0, 5, 10, 10, 10, 10, 10, 0, 10, 5, 10,
10, 10, 5, 10, 10, 10, 10, 10, 5, 10, 10, 10, 10, 5, 5, 10,
5, 10, 0, 10, 10, 5, 5, 10, 5, 10, 10, 10, 10), ps_a4_1 = c(10,
10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10,
10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10,
10, 10, 5, 5, 10, 5, 10, 10, 10, 10), ps_a4_2 = c(0, 10,
10, 10, 5, 10, 5, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10,
10, 5, 10, 5, 10, 10, 10, 5, 10, 10, 10, 5, 0, 10, 10, 10,
5, 0, 10, 5, 10, 10, 10, 10), ps_a4_3 = c(10, 0, 10, 5, 5,
10, 5, 10, 10, 5, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10,
5, 10, 10, 10, 5, 10, 10, 10, 5, 10, 10, 10, 10, 5, 0, 5,
0, 10, 5, 10, 10), ps_a4_4 = c(10, 10, 10, 10, 5, 10, 5,
10, 10, 0, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 5, 10,
10, 10, 10, 10, 10, 10, 5, 10, 5, 10, 10, 10, 10, 5, 5, 10,
10, 10, 10), ps_a4_5 = c(5, 5, 10, 5, 10, 5, 10, 10, 0, 0,
10, 10, 5, 10, 10, 10, 10, 10, 0, 10, 5, 5, 5, 10, 0, 10,
5, 10, 5, 0, 10, 10, 10, 10, 0, 5, 0, 5, 10, 10, 5), ps_a4_6 = c(5,
5, 0, 5, 0, 10, 0, 10, 5, 5, 10, 10, 5, 10, 10, 10, 0, 10,
5, 10, 5, 0, 5, 10, 5, 10, 5, 0, 5, 10, 0, 0, 10, 5, 0, 5,
0, 10, 10, 10, 10)), row.names = c(NA, -41L), class = "data.frame")
CodePudding user response:
I followed your idea of pivoting longer, using pivot_longer() from tidyr to put the scale groups in rows but leave the items in columns. (The last two examples in the documentation for pivot_longer() are my go-to when trying to remember how to do this.)
However, this relies on you having the same number of items for each scale; I'm not sure how it will hold up for varying items per scale.
Once things are in a longer form, use a nest_by() on quest and the scales variable followed by mutate() to nest and calculate the alpha for each row.
I didn't paste all the warnings and messages here, but there were loads. You can also remove the data column at the end if you don't need it any longer.
library(psych)
library(dplyr)
library(tidyr)
d %>%
pivot_longer(cols = -quest,
names_to = c("scale", ".value"),
names_pattern = "(\\w _\\w _)(.)") %>%
nest_by(quest, scale) %>%
mutate(alpha(data)$total)
#> # A tibble: 15 x 12
#> # Rowwise: quest, name
#> quest name data raw_alpha std.alpha `G6(smc)` average_r `S/N` ase
#> <dbl> <chr> <list<t> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 2 cg_a4_ [16 x 6] 0.619 0.594 0.728 0.226 1.46 0.141
#> 2 2 com_a~ [16 x 6] 0.810 0.808 0.881 0.412 4.20 0.0719
#> 3 2 fm_a4_ [16 x 6] 0.400 0.421 0.546 0.108 0.728 0.221
#> 4 2 gm_a4_ [16 x 6] 0.842 0.952 0.745 0.831 19.7 0.0592
#> 5 2 ps_a4_ [16 x 6] 0.684 0.753 0.870 0.337 3.05 0.123
#> 6 4 cg_a4_ [15 x 6] 0.677 0.696 0.807 0.276 2.29 0.126
#> 7 4 com_a~ [15 x 6] 0.673 0.613 0.842 0.209 1.58 0.110
#> 8 4 fm_a4_ [15 x 6] 0.669 0.714 0.811 0.294 2.50 0.124
#> 9 4 gm_a4_ [15 x 6] 0.811 0.759 0.873 0.386 3.15 0.0389
#> 10 4 ps_a4_ [15 x 6] 0.533 0.551 0.605 0.170 1.23 0.161
#> 11 6 cg_a4_ [10 x 6] -0.168 -0.00601 0.550 -0.00120 -0.00597 0.621
#> 12 6 com_a~ [10 x 6] -0.184 0.228 0.486 0.0686 0.295 0.644
#> 13 6 fm_a4_ [10 x 6] 0.508 0.542 0.727 0.191 1.18 0.248
#> 14 6 gm_a4_ [10 x 6] -0.075 -0.492 -0.0806 -0.0582 -0.330 0.398
#> 15 6 ps_a4_ [10 x 6] 0.844 0.879 0.903 0.592 7.26 0.0710
#> # ... with 3 more variables: mean <dbl>, sd <dbl>, median_r <dbl>
Created on 2021-09-23 by the reprex package (v2.0.0)
CodePudding user response:
Feels like grouping/pivoting is trying to over-engineer a solution. One approach could be to write a function that allows you to set the value that sits within contains().
library(psych)
library(tidyverse)
apply_alpha <- function(data, nest_contains) {
data %>%
select(quest, contains(nest_contains)) %>%
group_by(quest) %>%
do(alpha(.)$total)
}
apply_alpha(d, 'com_')
apply_alpha(d, 'gm_')
apply_alpha(d, 'fm_')
Important to note that with this approach I'm receiving a large number of warning messages that I'm not familiar with. They're coming from the use of the alpha() function.
CodePudding user response:
You could to something like this: Let me know to explain if this is what you are looking for:
library(tidyverse)
library(psych)
reg_fm_a4_ <- "^fm_a4_.*"
reg_com_a4_ <- "^com_a4_.*"
reg_gm_a4_ <- "^gm_a4_.*"
reg_cg_a4_ <- "^cg_a4_.*"
reg_ps_a4_ <- "^ps_a4_.*"
regs <- c(reg_fm_a4_, reg_com_a4_, reg_gm_a4_, reg_cg_a4_, reg_ps_a4_) %>%
set_names(c("fm_a4_", "com_a4_", "gm_a4_", "cg_a4_",
"ps_a4_"))
cronbachs_alpha <-
map_df(regs, ~
d %>%
select(dplyr::matches(.x)) %>%
psych::alpha(check.keys = TRUE) %>% .$total %>%
tibble::rownames_to_column()
,.id = "scale"
)
scale rowname raw_alpha std.alpha G6(smc) average_r S/N ase mean sd median_r
1 fm_a4_ 0.4655172 0.4841889 0.5081686 0.1352840 0.9386944 0.12722395 8.008130 1.716728 0.16047102
2 com_a4_ 0.7246145 0.7294824 0.7367755 0.3100766 2.6966174 0.06440378 8.130081 2.056329 0.32419199
3 gm_a4_ 0.6285083 0.6818823 0.7360522 0.2632152 2.1434909 0.08701602 7.337398 1.516341 0.09958706
4 cg_a4_ 0.5260735 0.5134628 0.5966499 0.1495805 1.0553414 0.10814655 6.524390 1.737080 0.12196703
5 ps_a4_ 0.7173328 0.7486200 0.7597498 0.3317028 2.9780417 0.06479382 7.906504 1.990620 0.36281243
CodePudding user response:
You could do your reshaping and then work with nested data. Of course, if you don't want to keep the nested data in your results, you can just unselect the data column.
The advantage of this solution (if you see it as such) is that you a) don't need to create extra objects nor b) specific functions.
d %>%
mutate(id = 1:n()) %>%
pivot_longer(cols = c(-id, -quest)) %>%
separate(col = name,
into = c("scale", "item"),
sep = "_",
extra = "merge") %>%
pivot_wider(names_from = item) %>%
select(-id) %>%
group_by(quest, scale) %>%
nest() %>%
mutate(alpha_results = map(data, ~alpha(.)$total)) %>%
unnest_wider(alpha_results) %>%
arrange(scale, quest)
which gives:
# Groups: quest, scale [15]
quest scale data raw_alpha std.alpha `G6(smc)` average_r `S/N` ase mean sd median_r
<dbl> <chr> <list> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 2 cg <tibble [16 x 6]> 0.619 0.594 0.728 0.226 1.46 0.141 7.60 1.90 0.157
2 4 cg <tibble [15 x 6]> 0.677 0.696 0.807 0.276 2.29 0.126 8 1.88 0.368
3 6 cg <tibble [10 x 6]> -0.168 -0.00601 0.550 -0.00120 -0.00597 0.621 8.42 1.07 0.102
4 2 com <tibble [16 x 6]> 0.810 0.808 0.881 0.412 4.20 0.0719 7.24 2.63 0.457
5 4 com <tibble [15 x 6]> 0.673 0.613 0.842 0.209 1.58 0.110 8.83 1.60 0.201
6 6 com <tibble [10 x 6]> -0.184 0.228 0.486 0.0686 0.295 0.644 8.5 0.946 0.0970
7 2 fm <tibble [16 x 6]> 0.4 0.421 0.546 0.108 0.728 0.221 8.12 1.62 0.0953
8 4 fm <tibble [15 x 6]> 0.669 0.714 0.811 0.294 2.50 0.124 8.28 1.82 0.366
9 6 fm <tibble [10 x 6]> 0.508 0.542 0.727 0.191 1.18 0.248 7.42 1.73 0.279
10 2 gm <tibble [16 x 6]> 0.842 0.952 0.745 0.831 19.7 0.0592 9.48 1.05 0.831
11 4 gm <tibble [15 x 6]> 0.811 0.759 0.873 0.386 3.15 0.0389 8.83 1.94 0.511
12 6 gm <tibble [10 x 6]> -0.075 -0.492 -0.0806 -0.0582 -0.330 0.398 8.33 1.11 -0.111
13 2 ps <tibble [16 x 6]> 0.684 0.753 0.870 0.337 3.05 0.123 7.08 1.97 0.316
14 4 ps <tibble [15 x 6]> 0.533 0.551 0.605 0.170 1.23 0.161 8.83 1.29 0.150
15 6 ps <tibble [10 x 6]> 0.844 0.879 0.903 0.592 7.26 0.0710 7.83 2.43 0.604