I have used a random forest for predicting classes. Now, I am trying to plot variable importance for each class. I have used the below code, but it does not provide me varImp class wise, it is giving me for whole model. Can someone please help me.
Thank you.
odFit = train(x = df_5[,-22],
y = df_5$`kpres$cluster`,
ntree=20,method="rf",metric = "Accuracy",trControl = control,tuneGrid = tunegrid
)
odFit
varImp(odFit)
CodePudding user response:
Just add importance=TRUE in the train function, which is the same to do importance(odFit) in the randomForest package.
Here a reproducible example:
library(caret)
data(iris)
control <- trainControl(method = "cv",10)
tunegrid <- expand.grid(mtry=2:ncol(iris)-1)
odFit = train(x = iris[,-5],
y = iris$Species,
ntree=20,
trControl = control,
tuneGrid = tunegrid,
importance=T
)
odFit
varImp(odFit)
and here is the output
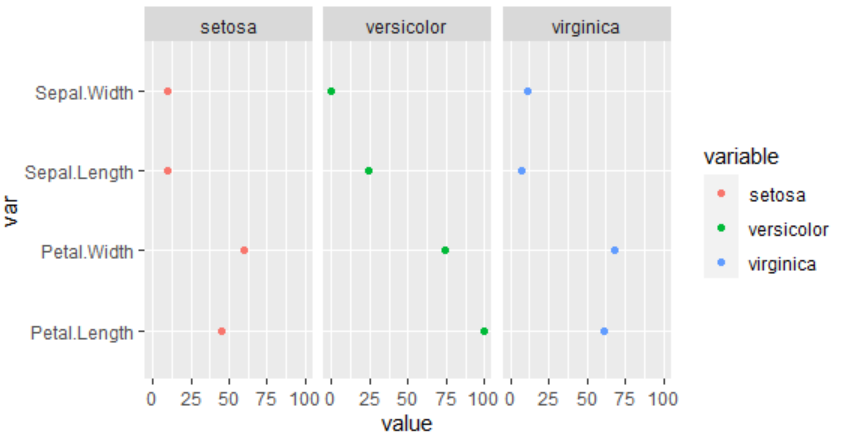
rf variable importance
variables are sorted by maximum importance across the classes
setosa versicolor virginica
Petal.Width 57.21 73.747 100.00
Petal.Length 61.90 79.981 77.49
Sepal.Length 20.01 2.867 40.47
Sepal.Width 20.01 0.000 15.73
you can plot the variable importance with ggplot
library(ggplot2)
vi <- varImp(odFit,scale=T)[[1]]
vi$var <-row.names(vi)
vi <- reshape2::melt(vi)
ggplot(vi,aes(value,var,col=variable))
geom_point()
facet_wrap(~variable)