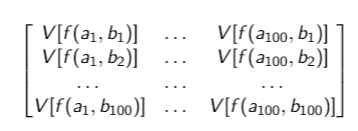I need to have a more efficient code, that the one implemented by me below. Is it possible to have a more efficient code, also using numpy?
I am going to explain my algorithm and how it works. Considering the figure below, I have a matrix that contains 10000 matrix inside, and each matrix inside has a dimension 100x100.
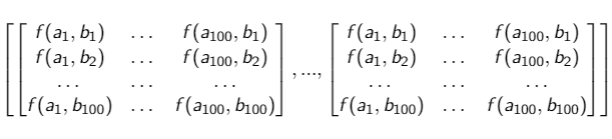
My output should be a matrix 100x100, like this:
therefore, the first element of my "output" V[f(a_1,b_1)] is the variance of all f(a_1,b_1) elements contained in 10000 matrices. Thus, I need to compute the variances of the elements with the same index.
Below there is the code used, it should be correct (I hope..). is it possible to have a more efficient code? (for simplicity, I simulate the input with random numbers, since the matrix is too big to post here)
import numpy as np
input = np.random.randint(0, 100, size=(10000, 100, 100))
output = []
for n in range(100):
row = []
for i in range(100):
row2 = []
for m in range(10000):
P_mi_sigmai = input[m][n][i]
row2.append(P_mi_sigmai)
variance = np.var(row2)
row.append(variance)
output.append(row)
print(np.array(output).shape)
CodePudding user response:
Here's how i would do it:
output = input.var(axis = 0)
Here's a minimal reprex:
import numpy as np
arr = np.random.rand(5, 2, 2)
arr[:, 0, 0] = 1
var_arr = arr.var(0)
print(f'arr = \n{arr}')
print(f'var_arr = \n{var_arr}')
output:
arr =
[[[1. 0.13682225]
[0.24076008 0.61107865]]
[[1. 0.99948733]
[0.61871626 0.64518322]]
[[1. 0.7979549 ]
[0.53991881 0.17229415]]
[[1. 0.13547922]
[0.97390205 0.50778721]]
[[1. 0.74116566]
[0.05428085 0.86287107]]]
var_arr =
[[0. 0.12837323]
[0.10101842 0.05092759]]
I'm not gonna show it here, but using your code i get the same result