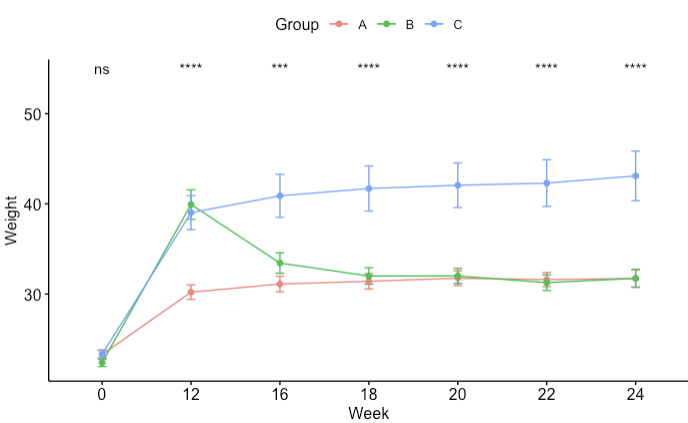
I'm relatively new to programming, so bear with me. I'm analysing some biological data, where the weight of mice have been measured across several weeks. Mice were allocated into 3 treatment groups: A, B and C. A is the reference group. What I want, is a plot showing the statistical comparisons of weight for each week, preferably with different significance symbols for each group. From the data it is clear, that the body weight of group B normalizes after 12 weeks, whereas group C stays high. I want to be able to see that statistical change on the plot. Hope that makes sense. For some reason I just can't figure it out. Here's my code, and how the plot looks
BW %>%
gather(Week, Weight, "0":"24") %>%
group_by(Group, Week) %>%
ggline(x = "Week", y = "Weight", add = "mean_se", color = "Group")
stat_compare_means(aes(group = Group), label = "p.signif", method = "anova")
CodePudding user response:
library(tidyverse)
library(ggpubr)
library(broom)
# create example data
set.seed(1337)
base_values <- tribble(
~Group, ~Week, ~base_value,
"A", 0, 20,
"A", 12, 30,
"A", 16, 32,
"B", 0, 21,
"B", 12, 35,
"B", 16, 32,
"C", 0, 20,
"C", 12, 38,
"C", 16, 40
)
data <-
tibble(Group = LETTERS[1:3]) %>%
expand_grid(Week = c(0, 12, 16)) %>%
left_join(base_values) %>%
mutate(Weight = base_value %>% map(~ runif(n = 10, min = 0.8, max = 1.2) * .x)) %>%
select(-base_value) %>%
unnest(Weight)
#> Joining, by = c("Group", "Week")
data
#> # A tibble: 90 x 3
#> Group Week Weight
#> <chr> <dbl> <dbl>
#> 1 A 0 20.6
#> 2 A 0 20.5
#> 3 A 0 16.6
#> 4 A 0 19.6
#> 5 A 0 19.0
#> 6 A 0 18.7
#> 7 A 0 23.6
#> 8 A 0 18.2
#> 9 A 0 18.0
#> 10 A 0 17.2
#> # … with 80 more rows
tests <-
expand_grid(
Group = c("B", "C"),
Week = c(0, 12, 16)
) %>%
mutate(
test = list(Group, Week) %>% pmap(~ {
data2 <- data %>%
filter(Week == .y) %>%
pivot_wider(names_from = Group, values_from = Weight)
wilcox.test(data2[["A"]][[1]], data2[[.x]][[1]]) %>% tidy()
})
) %>%
unnest(test) %>%
mutate(
label = case_when(p.value < 0.01 ~ "**", p.value < 0.05 ~ "*", TRUE ~ "n.s.")
) %>%
left_join(
# position of labels
data %>%
group_by(Group, Week) %>%
summarise(Weight = mean(Weight) - 30)
)
#> Warning: Values are not uniquely identified; output will contain list-cols.
#> * Use `values_fn = list` to suppress this warning.
#> * Use `values_fn = length` to identify where the duplicates arise
#> * Use `values_fn = {summary_fun}` to summarise duplicates
#> Warning: Values are not uniquely identified; output will contain list-cols.
#> * Use `values_fn = list` to suppress this warning.
#> * Use `values_fn = length` to identify where the duplicates arise
#> * Use `values_fn = {summary_fun}` to summarise duplicates
#> Warning: Values are not uniquely identified; output will contain list-cols.
#> * Use `values_fn = list` to suppress this warning.
#> * Use `values_fn = length` to identify where the duplicates arise
#> * Use `values_fn = {summary_fun}` to summarise duplicates
#> Warning: Values are not uniquely identified; output will contain list-cols.
#> * Use `values_fn = list` to suppress this warning.
#> * Use `values_fn = length` to identify where the duplicates arise
#> * Use `values_fn = {summary_fun}` to summarise duplicates
#> Warning: Values are not uniquely identified; output will contain list-cols.
#> * Use `values_fn = list` to suppress this warning.
#> * Use `values_fn = length` to identify where the duplicates arise
#> * Use `values_fn = {summary_fun}` to summarise duplicates
#> Warning: Values are not uniquely identified; output will contain list-cols.
#> * Use `values_fn = list` to suppress this warning.
#> * Use `values_fn = length` to identify where the duplicates arise
#> * Use `values_fn = {summary_fun}` to summarise duplicates
#> `summarise()` has grouped output by 'Group'. You can override using the `.groups` argument.
#> Joining, by = c("Group", "Week")
tests
#> # A tibble: 6 x 8
#> Group Week statistic p.value method alternative label Weight
#> <chr> <dbl> <dbl> <dbl> <chr> <chr> <chr> <dbl>
#> 1 B 0 27 0.0892 Wilcoxon rank sum exac… two.sided n.s. -8.48
#> 2 B 12 38 0.393 Wilcoxon rank sum exac… two.sided n.s. 3.93
#> 3 B 16 63 0.353 Wilcoxon rank sum exac… two.sided n.s. 2.86
#> 4 C 0 42 0.579 Wilcoxon rank sum exac… two.sided n.s. -9.58
#> 5 C 12 22 0.0355 Wilcoxon rank sum exac… two.sided * 6.58
#> 6 C 16 17 0.0115 Wilcoxon rank sum exac… two.sided * 11.3
data %>%
ggline(x = "Week", y = "Weight", add = "mean_se", color = "Group")
geom_text(
data = tests %>% mutate(Week = Week %>% factor()),
mapping = aes(label = label, color = Group, y = 45),
size = 10
)
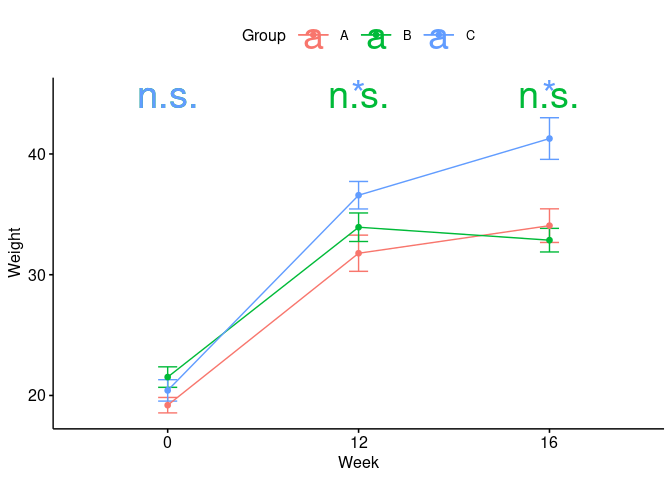
Created on 2021-12-01 by the reprex package (v2.0.1)