i have a data frame that looks like this :
total-36 columns.
data sample code :
df <-
structure(
list(
Bacteroidaceae = c(
0,
0.10944999,
0.104713314,
0.125727668,
0.124136247,
0.005155911,
0.005072778,
0.010231826,
0.010188139
),
Christensenellaceae = c(
0,
0.009910731,
0.010131195,
0.009679938,
0.01147601,
0.010484508,
0.008641566,
0.010017172,
0.010741488
),
treatment = c(
"Original Sample1",
"Original Sample2",
"Original Sample3",
"Original Sample4",
"treatment1_1",
"treatment1_2",
"treatment1_3",
"treatment1_4"
)
),
class = "data.frame",
row.names = c(NA,-8L)
)
what i wish to do is to creat plots for all the columns in the data so evey column would have 2 plots : one for treatment1 and one for the original sample in total 72 plots
for example:
same for the original sample type i tried using this code:
df %>%
tidyr::pivot_longer(!treatment, names_to = "taxa", values_to = "value") %>%
dplyr::filter(str_detect(treatment, "Treatment1")) %>%
for (i in columns(df)){
)
ggplot(aes(x = treatment, y = value, color = taxa),group=treatment)
geom_point()
stat_summary(fun.y = mean,
geom = "line", width = 0.5) geom_jitter(widh=0.25)
theme_bw()}}
but it didnt work . is there any other way ?
thank you
CodePudding user response:
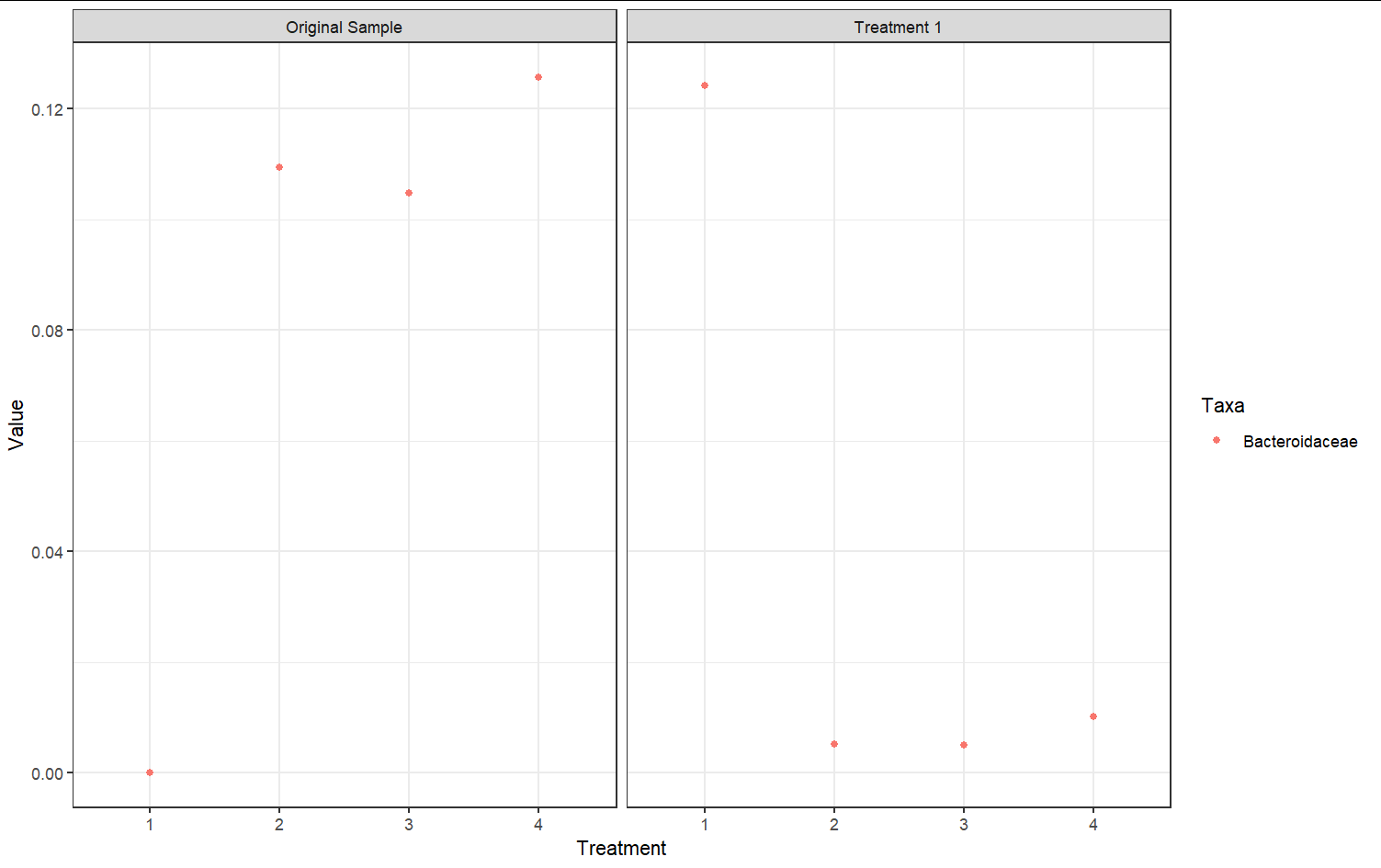
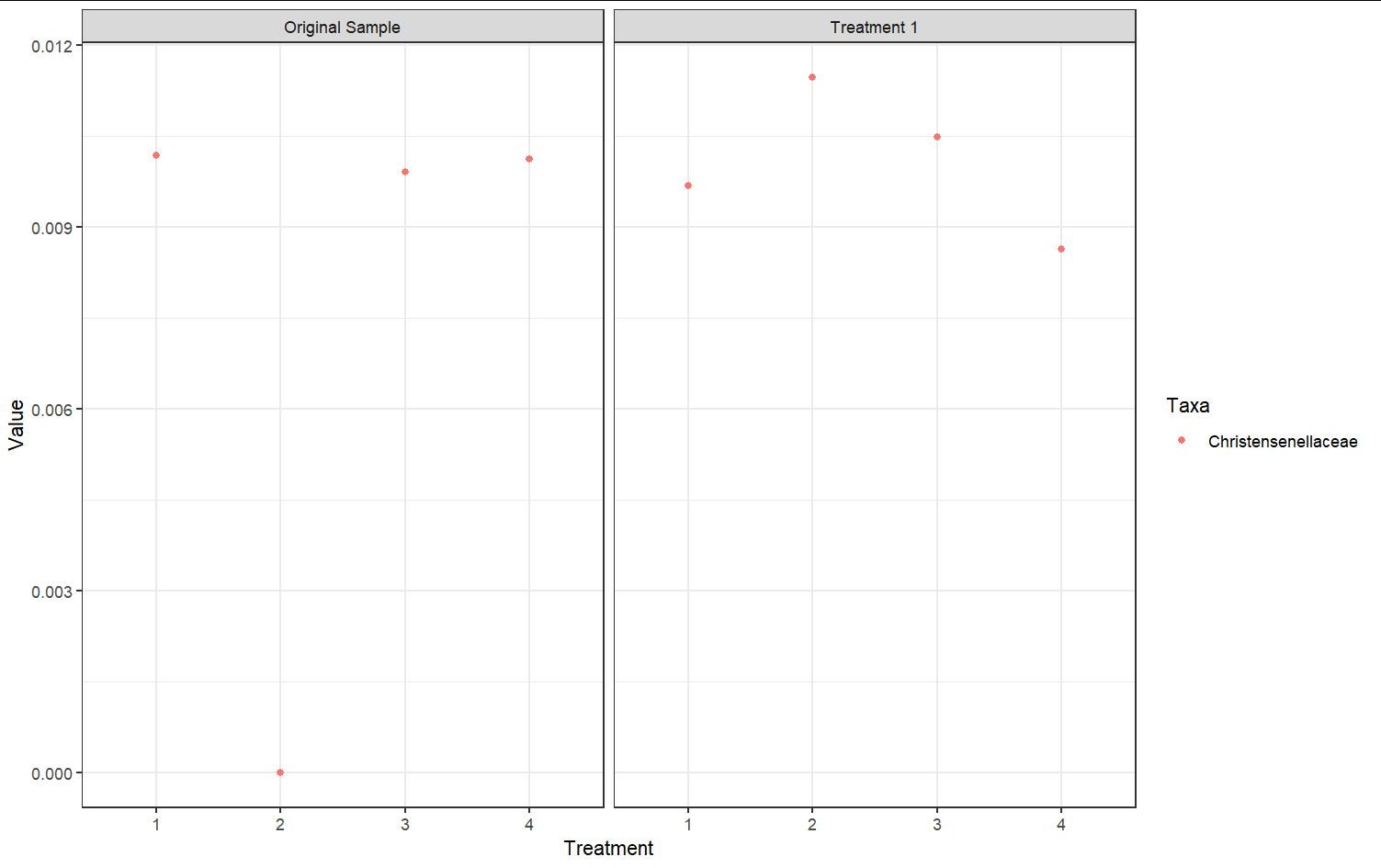
Perhaps this is what you are looking for:
library(tidyverse)
df %>%
pivot_longer(-treatment) %>%
mutate(plot = ifelse(str_detect(treatment, "Original"),
"Original Sample",
"Treatment 1"),
treatment = str_extract(treatment, "\\d $")) %>%
group_by(name) %>%
group_split() %>%
map(~.x %>% ggplot(aes(x = factor(treatment), y = value, color = factor(name)))
geom_point()
facet_wrap(~plot)
labs(x = "Treatment", y = "Value", color = "Taxa")
theme_bw())
This produces two plots (based on the test data):