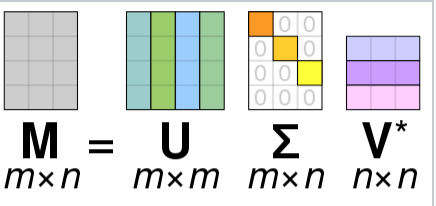
And this is what you get back in scipy.linalg.svd:
Factorizes the matrix a into two unitary matrices U and Vh, and a 1-D array s of singular values (real, non-negative) such that a == U @ S @ Vh, where S is a suitably shaped matrix of zeros with main diagonal s.
Whereas for svd in R they return you V. Therefore should be:
Vt = t(internal$v)
Then in R:
ZCA_svd(x)
head(ZCA_svd(x))
[,1] [,2] [,3] [,4] [,5] [,6]
[1,] 0.26067006 0.02112997 0.09365719 0.01843731 0.05470893 0.01750415
[2,] -0.17174605 -0.23530453 0.15122167 -0.27738192 0.03830312 -0.21142466
[3,] -0.10659408 0.07042392 0.06732517 -0.12081178 0.09487670 -0.01726953
[4,] 0.10659431 0.13668984 0.18523379 0.03799714 0.06525643 -0.09888497
[5,] -0.12998931 -0.05254591 -0.14654516 -0.15600721 0.13455552 -0.09930468
[6,] -0.07010493 0.01084335 -0.05152612 -0.07803706 -0.03505320 0.43416503
[,7] [,8] [,9]
[1,] -0.02021101 0.08766270 0.073049749
[2,] 0.15877490 0.24157032 0.009806777
[3,] 0.03148085 0.09361557 0.100372380
[4,] -0.03620529 0.09898168 0.044607751
[5,] 0.02847737 -0.30396604 0.574410291
[6,] 0.03105272 0.13842155 0.076071540
In python:
pd.DataFrame(svd_whiten(df.iloc[:,2:])).head(6)
0 1 2 3 4 5 6 7 8
0 0.260670 0.021130 0.093657 0.018437 0.054709 0.017504 -0.020211 0.087663 0.073050
1 -0.171746 -0.235305 0.151222 -0.277382 0.038303 -0.211425 0.158775 0.241570 0.009807
2 -0.106594 0.070424 0.067325 -0.120812 0.094877 -0.017270 0.031481 0.093616 0.100372
3 0.106594 0.136690 0.185234 0.037997 0.065256 -0.098885 -0.036205 0.098982 0.044608
4 -0.129989 -0.052546 -0.146545 -0.156007 0.134556 -0.099305 0.028477 -0.303966 0.574410
5 -0.070105 0.010843 -0.051526 -0.078037 -0.035053 0.434165 0.031053 0.138422 0.076072