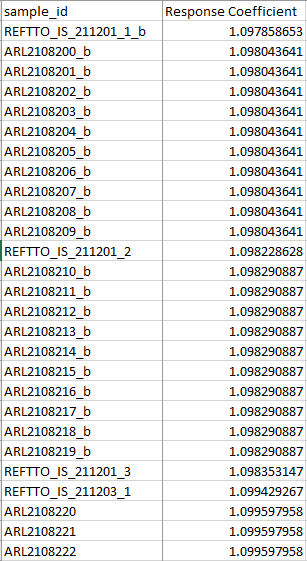
I have a data set that has measured values in some samples and leaves NAs in others. In between each measured value is 10 samples with no value. I want the value of those ten samples to each be the average of the proceeding and following measured values. The data looks like this:
Importantly, the values for each of the ten NAs between any two known values has to be the same. I previously tried using zoo::na.approx(RC, rule=2), but that calculates averages using its own interpolated data, not just the two known values. The output should look like this:
Edit as per deschen's request:
dput(rc_report[,c(2,26)]) structure(list(sample_id = c("REFTTO_IS_211201_1_b", "ARL2108200_b", "ARL2108201_b", "ARL2108202_b", "ARL2108203_b", "ARL2108204_b", "ARL2108205_b", "ARL2108206_b", "ARL2108207_b", "ARL2108208_b", "ARL2108209_b", "REFTTO_IS_211201_2", "ARL2108210_b", "ARL2108211_b", "ARL2108212_b", "ARL2108213_b", "ARL2108214_b", "ARL2108215_b", "ARL2108216_b", "ARL2108217_b", "ARL2108218_b", "ARL2108219_b", "REFTTO_IS_211201_3", "REFTTO_IS_211203_1", "ARL2108220", "ARL2108221", "ARL2108222", "ARL2108223", "ARL2108224", "ARL2108225", "ARL2108226", "ARL2108227", "ARL2108228", "ARL2108229", "REFTTO_IS_211203_2", "ARL2108230", "ARL2108231", "ARL2108232", "ARL2108233", "ARL2108234", "ARL2108235", "ARL2108236", "ARL2108237", "ARL2108238", "ARL2108239", "REFTTO_IS_211203_3", "REFTTO_IS_211206_1", "ARL2108240", "ARL2108241", "ARL2108242", "ARL2108243", "ARL2108244", "ARL2108245", "ARL2108246", "ARL2108247", "ARL2108248", "ARL2108249", "REFTTO_IS_211206_2", "ARL2108250", "ARL2108251", "ARL2108252", "ARL2108253", "ARL2108254", "ARL2108255", "ARL2108256", "ARL2108258", "ARL2108259", "REFTTO_IS_211206_3" ), response_coefficient = c("1.09785865302384", "NA", "NA", "NA", "NA", "NA", "NA", "NA", "NA", "NA", "NA", "1.09822862814289", "NA", "NA", "NA", "NA", "NA", "NA", "NA", "NA", "NA", "NA", "1.09835314677401", "1.09942926690693", "NA", "NA", "NA", "NA", "NA", "NA", "NA", "NA", "NA", "NA", "1.10084276861106", "NA", "NA", "NA", "NA", "NA", "NA", "NA", "NA", "NA", "NA", "1.10078178211056", "1.11104600880183", "NA", "NA", "NA", "NA", "NA", "NA", "NA", "NA", "NA", "NA", "1.11203467893562", "NA", "NA", "NA", "NA", "NA", "NA", "NA", "NA", "NA", "1.11344223852824" )), class = "data.frame", row.names = c(NA, -68L))
CodePudding user response:
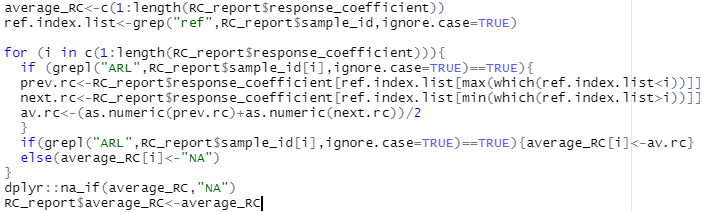
I solved this problem using the following code. It's probably pretty inefficient, but it does the job.
CodePudding user response:
Using the data shown in the Note at the end (we changed the column names to shorter ones) use na.locf0 both forwards and backwards taking the average.
library(dplyr)
library(zoo)
dd %>%
mutate(coef = as.numeric(ifelse(coef == "NA", NA, coef)),
coef = (na.locf0(coef) na.locf0(coef, fromLast = TRUE)) / 2)
Note
dd <- structure(list(sample_id = c("REFTTO_IS_211201_1_b", "ARL2108200_b", "ARL2108201_b", "ARL2108202_b", "ARL2108203_b", "ARL2108204_b", "ARL2108205_b", "ARL2108206_b", "ARL2108207_b", "ARL2108208_b", "ARL2108209_b", "REFTTO_IS_211201_2", "ARL2108210_b", "ARL2108211_b", "ARL2108212_b", "ARL2108213_b", "ARL2108214_b", "ARL2108215_b", "ARL2108216_b", "ARL2108217_b", "ARL2108218_b", "ARL2108219_b", "REFTTO_IS_211201_3", "REFTTO_IS_211203_1", "ARL2108220", "ARL2108221", "ARL2108222", "ARL2108223", "ARL2108224", "ARL2108225", "ARL2108226", "ARL2108227", "ARL2108228", "ARL2108229", "REFTTO_IS_211203_2", "ARL2108230", "ARL2108231", "ARL2108232", "ARL2108233", "ARL2108234", "ARL2108235", "ARL2108236", "ARL2108237", "ARL2108238", "ARL2108239", "REFTTO_IS_211203_3", "REFTTO_IS_211206_1", "ARL2108240", "ARL2108241", "ARL2108242", "ARL2108243", "ARL2108244", "ARL2108245", "ARL2108246", "ARL2108247", "ARL2108248", "ARL2108249", "REFTTO_IS_211206_2", "ARL2108250", "ARL2108251", "ARL2108252", "ARL2108253",
"ARL2108254", "ARL2108255", "ARL2108256", "ARL2108258", "ARL2108259", "REFTTO_IS_211206_3" ),
response_coefficient = c("1.09785865302384", "NA", "NA", "NA", "NA", "NA",
"NA", "NA", "NA", "NA", "NA", "1.09822862814289", "NA", "NA", "NA", "NA",
"NA", "NA", "NA", "NA", "NA", "NA", "1.09835314677401", "1.09942926690693",
"NA", "NA", "NA", "NA", "NA", "NA", "NA", "NA", "NA", "NA",
"1.10084276861106", "NA", "NA", "NA", "NA",
"NA", "NA", "NA", "NA", "NA", "NA", "1.10078178211056", "1.11104600880183",
"NA", "NA", "NA", "NA", "NA", "NA", "NA", "NA", "NA", "NA", "1.11203467893562",
"NA", "NA", "NA", "NA", "NA", "NA", "NA", "NA", "NA", "1.11344223852824" )),
class = "data.frame", row.names = c(NA, -68L))
names(dd) <- c("id", "coef")