Let's say I have a dataframe like below,
0 1 2 3 4
0 (989, 998) (1074, 999) (1159, 1000) (1244, 1001) (1329, 1002)
1 (970, 1042) (1057, 1043) (1143, 1044) (1230, 1045) (1316, 1046)
2 (951, 1088) (1039, 1089) (1127, 1090) (1214, 1091) (1302, 1092)
3 (930, 1137) (1020, 1138) (1109, 1139) (1198, 1140) (1287, 1141)
4 (909, 1188) (1000, 1189) (1091, 1190) (1181, 1191) (1271, 1192)
Each cell has x and y coordinates in tuple. I have an input called I and that's also x and Y coordinate in tuple. My goal is find nearest point for the input I.
Sample Input:
(1080, 1000)
Sample Output:
(1074, 999)
I have tried below snippet.
def find_nearest(array, key):
min_ = 1000
a = 0
b = 0
for item in array:
diff = abs(item[0]-key[0]) abs(item[1]-key[1])
if diff<min_:
min_ = diff
a,b = item
if diff==0:
return (a,b)
return (a,b)
find_nearest(sum(df.values.tolist(), []), I)
This gives me what I expected. But, Is there any efficient solution for the problem?
CodePudding user response:
Try:
# Setup
data = [[(989, 998), (1074, 999), (1159, 1000), (1244, 1001), (1329, 1002)],
[(970, 1042), (1057, 1043), (1143, 1044), (1230, 1045), (1316, 1046)],
[(951, 1088), (1039, 1089), (1127, 1090), (1214, 1091), (1302, 1092)],
[(930, 1137), (1020, 1138), (1109, 1139), (1198, 1140), (1287, 1141)],
[(909, 1188), (1000, 1189), (1091, 1190), (1181, 1191), (1271, 1192)]]
df = pd.DataFrame(data)
l = (1080, 1000)
out = min(df.to_numpy().flatten(), key=lambda c: (c[0]- l[0])**2 (c[1]-l[1])**2)
print(out)
# Output:
(1074, 999)
Update:
Is there any way, I can get df index of nearest element?
dist = df.stack().apply(lambda c: (c[0]- l[0])**2 (c[1]-l[1])**2)
idx = dist.index[dist.argmin()]
val = df.loc[idx]
print(idx)
print(val)
# Output:
(0, 1)
(1074, 999)
Update 2
But, Is there any efficient solution for the problem?
arr = df.to_numpy().astype([('x', int), ('y', int)])
dist = (arr['x'] - l[0])**2 (arr['y'] - l[1])**2
idx = tuple(np.argwhere(dist == np.min(dist))[0])
val = arr[idx] # or df.loc[idx]
CodePudding user response:
How about this snippet I wrote?
# cordinates: np.ndarray(n, 2)
def find_nearest(cordinates, x, y):
x_d = np.abs(cordinate[:, 0] - x)
y_d = np.abs(cordinate[:, 1] - y)
nearest_idx = np.argmin(x_d y_d)
return cordinate[nearest_idx]
CodePudding user response:
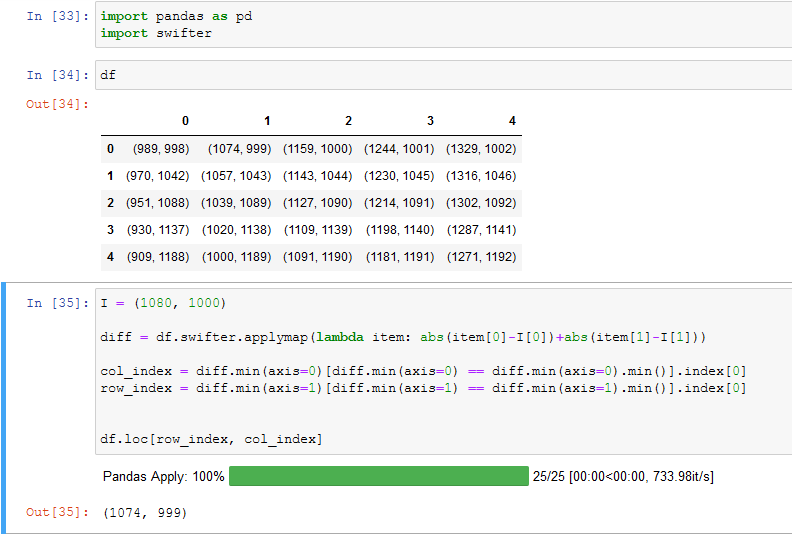
You can use swifter and applymap for faster processing
I = (1080, 1000)
diff = df.swifter.applymap(lambda item: abs(item[0]-I[0]) abs(item[1]-I[1]))
col_index = diff.min(axis=0)[diff.min(axis=0) == diff.min(axis=0).min()].index[0]
row_index = diff.min(axis=1)[diff.min(axis=1) == diff.min(axis=1).min()].index[0]
df.loc[row_index, col_index]
CodePudding user response:
It appears you just need a two-column DataFrame and find distance between each row and a sample coordinate. So here is my implementation:
Your data when copied came off as strings. You don't actually need this line:
data = pd.Series(df.to_numpy().flatten()).str.strip().str.strip('()').str.split(',', expand=True).astype(int)
sample = (1080, 1000)
Solution start here:
distances = data.apply(lambda x: (x[0]-sample[0])**2 (x[1]-sample[1])**2, axis=1)
out = tuple(data[distances == distances.min()].to_numpy()[0])
Output:
(1074, 999)
CodePudding user response:
Update 2
However, in a larger Dataframe, it starts to dropdown: