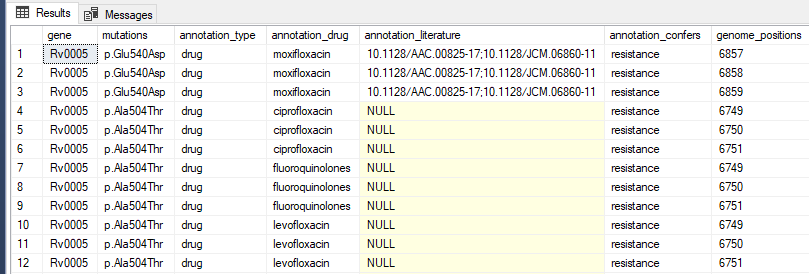
I am confused how to shred my JSON data into a table because is not formatted with names for the arrays
The actual JSON file is much bigger (19K lines) so I only pulled a small portion of it out (the first two of top level and a few from within those.
DECLARE @txt1 varchar(max) = '{ "Rv0005": { "p.Glu540Asp": { "annotations": [ { "type": "drug", "drug": "moxifloxacin", "literature": "10.1128/AAC.00825-17;10.1128/JCM.06860-11", "confers": "resistance" } ], "genome_positions": [ 6857, 6858, 6859 ] }, "p.Ala504Thr": { "annotations": [ { "type": "drug", "drug": "ciprofloxacin", "confers": "resistance" }, { "type": "drug", "drug": "fluoroquinolones", "confers": "resistance" }, { "type": "drug", "drug": "levofloxacin", "confers": "resistance" }, { "type": "drug", "drug": "moxifloxacin", "confers": "resistance" }, { "type": "drug", "drug": "ofloxacin", "confers": "resistance" } ], "genome_positions": [ 6749, 6750, 6751 ] }, "p.Ala504Val": { "annotations": [ { "type": "drug", "drug": "ciprofloxacin", "confers": "resistance" }, { "type": "drug", "drug": "fluoroquinolones", "confers": "resistance" }, { "type": "drug", "drug": "levofloxacin", "confers": "resistance" }, { "type": "drug", "drug": "moxifloxacin", "confers": "resistance" }, { "type": "drug", "drug": "ofloxacin", "confers": "resistance" } ], "genome_positions": [ 6749, 6750, 6751 ] } }, "Rv2043c": { "p.Thr100Ile": { "annotations": [ { "type": "drug", "drug": "pyrazinamide", "literature": "10.1128/JCM.01214-17", "confers": "resistance" } ], "genome_positions": [ 2288942, 2288943, 2288944 ] }, "p.Thr160Ala": { "annotations": [ { "type": "drug", "drug": "pyrazinamide", "literature": "10.1128/JCM.01214-17", "confers": "resistance" } ], "genome_positions": [ 2288762, 2288763, 2288764 ] }, "c.101_102insT": { "annotations": [ { "type": "drug", "drug": "pyrazinamide", "confers": "resistance" } ], "genome_positions": [ 2289140, 2289141 ] } } }'
SELECT * FROM OPENJSON(@txt1)
The top level is a gene and this is just the data from two genes (Rv0005 = gene 1, Rv2043c = gene 2). Each gene can have multiple mutations (e.g. Rv0005 has a mutation at p.Glu540Asp and p.Ala504Thr) and each of those mutations have some data associated with it (literature, resistance, genomic positions, etc.). I know I can parse portions of the JSON and JSON array out via
SELECT * FROM OPENJSON(@txt1)
SELECT * FROM OPENJSON(@txt1, '$.Rv0005."p.Glu540Asp".genome_positions')
But I don't know how to shred the whole thing out without knowing what the keys/values are. In particular there are 35 unique genes (the top of the JSON tree) and each of the mutations are named under them but are unique (e.g. p.Glu540Asp, etc).
Ultimately I'd either like to pull the data into multiple normalized tables but honestly one big table would be fine like this
CREATE TABLE #Muts (gene varchar(max), mutations varchar(max), annotation_type varchar(max), annotation_drug varchar(max), annotation_literature varchar(max), annotation_confers varchar(max), genome_positions int )
and the data for the first couple of values would look like this (notice that some mutations confer resistance to multiple drugs)
| gene | mutations | annotation_type | annotation_drug | annotation_literature | annotation_confers | genome_positions |
|---|---|---|---|---|---|---|
| Rv0005 | p.Glu540Asp | drug | moxifloxacin | 10.1128/AAC.00825-17;10.1128/JCM.06860-11 | resistance | 6857 |
| Rv0005 | p.Glu540Asp | drug | moxifloxacin | 10.1128/AAC.00825-17;10.1128/JCM.06860-11 | resistance | 6858 |
| Rv0005 | p.Glu540Asp | drug | moxifloxacin | 10.1128/AAC.00825-17;10.1128/JCM.06860-11 | resistance | 6859 |
| Rv0005 | p.Ala504Thr | drug | ciprofloxacin | 10.1128/AAC.00825-17;10.1128/JCM.06860-11 | resistance | 6849 |
| Rv0005 | p.Ala504Thr | drug | fluoroquinolones | 10.1128/AAC.00825-17;10.1128/JCM.06860-11 | resistance | 6849 |
| Rv0005 | p.Ala504Thr | drug | levofloxacin | 10.1128/AAC.00825-17;10.1128/JCM.06860-11 | resistance | 6849 |
| Rv0005 | p.Ala504Thr | drug | moxifloxacin | 10.1128/AAC.00825-17;10.1128/JCM.06860-11 | resistance | 6849 |
| Rv0005 | p.Ala504Thr | drug | ofloxacin | 10.1128/AAC.00825-17;10.1128/JCM.06860-11 | resistance | 6849 |
| Rv0005 | p.Ala504Thr | drug | ciprofloxacin | 10.1128/AAC.00825-17;10.1128/JCM.06860-11 | resistance | 6850 |
| Rv0005 | p.Ala504Thr | drug | fluoroquinolones | 10.1128/AAC.00825-17;10.1128/JCM.06860-11 | resistance | 6850 |
| Rv0005 | p.Ala504Thr | drug | levofloxacin | 10.1128/AAC.00825-17;10.1128/JCM.06860-11 | resistance | 6850 |
| Rv0005 | p.Ala504Thr | drug | moxifloxacin | 10.1128/AAC.00825-17;10.1128/JCM.06860-11 | resistance | 6850 |
| Rv0005 | p.Ala504Thr | drug | ofloxacin | 10.1128/AAC.00825-17;10.1128/JCM.06860-11 | resistance | 6850 |
| Rv0005 | p.Ala504Thr | drug | ciprofloxacin | 10.1128/AAC.00825-17;10.1128/JCM.06860-11 | resistance | 6851 |
| Rv0005 | p.Ala504Thr | drug | fluoroquinolones | 10.1128/AAC.00825-17;10.1128/JCM.06860-11 | resistance | 6851 |
| Rv0005 | p.Ala504Thr | drug | levofloxacin | 10.1128/AAC.00825-17;10.1128/JCM.06860-11 | resistance | 6851 |
| Rv0005 | p.Ala504Thr | drug | moxifloxacin | 10.1128/AAC.00825-17;10.1128/JCM.06860-11 | resistance | 6851 |
| Rv0005 | p.Ala504Thr | drug | ofloxacin | 10.1128/AAC.00825-17;10.1128/JCM.06860-11 | resistance | 6851 |
CodePudding user response:
You have to use CROSS APPLY with OPENJSON when you want to "pivot" a JSON array to a tabular table.
The following query return the expected result:
SELECT a.[key] as gene, b.[key] as mutations, c.*, d.value as genome_positions
FROM OPENJSON(@txt1) a
CROSS APPLY OPENJSON(a.value) b
CROSS APPLY OPENJSON(b.value,'$.annotations')
WITH (
annotation_type nvarchar(100) '$.type'
, annotation_drug nvarchar(100) '$.drug'
, annotation_literature nvarchar(100) '$.literature'
, annotation_confers nvarchar(100) '$.confers'
) c
CROSS APPLY OPENJSON(b.value,'$.genome_positions') d
Result:
CodePudding user response:
When 'type' is 5 the Value (of the kv pair) is an array. To get to the lowest level of the array you could try specifying the JSON schema along with OPENJSON.
/* specify explicity JSON schema */
/* to open bottom-most array */
select *
from openjson(@txt1) j
cross apply openjson(j.[value]) l1
cross apply openjson(l1.[value]) l2
cross apply openjson(l2.[value]) l3
cross apply openjson(l3.[value])
with ([type] nvarchar(4000),
drug nvarchar(4000),
literature nvarchar(4000),
confers nvarchar(4000))
where l3.[type]=5;
The rest of the leaf-level fields could be accessed by filtering the 'type' column.
/* open the rest of the fields */
select *
from openjson(@txt1) j
cross apply openjson(j.[value]) l1
cross apply openjson(l1.[value]) l2
cross apply openjson(l2.[value]) l3
where l3.[type]<>5;
CodePudding user response:
Please try the following solution.
SQL
DECLARE @json NVARCHAR(MAX) =
N'{
"Rv0005": {
"p.Glu540Asp": {
"annotations": [
{
"type": "drug",
"drug": "moxifloxacin",
"literature": "10.1128/AAC.00825-17;10.1128/JCM.06860-11",
"confers": "resistance"
}
],
"genome_positions": [
6857,
6858,
6859
]
},
"p.Ala504Thr": {
"annotations": [
{
"type": "drug",
"drug": "ciprofloxacin",
"confers": "resistance"
},
{
"type": "drug",
"drug": "fluoroquinolones",
"confers": "resistance"
},
{
"type": "drug",
"drug": "levofloxacin",
"confers": "resistance"
},
{
"type": "drug",
"drug": "moxifloxacin",
"confers": "resistance"
},
{
"type": "drug",
"drug": "ofloxacin",
"confers": "resistance"
}
],
"genome_positions": [
6749,
6750,
6751
]
},
"p.Ala504Val": {
"annotations": [
{
"type": "drug",
"drug": "ciprofloxacin",
"confers": "resistance"
},
{
"type": "drug",
"drug": "fluoroquinolones",
"confers": "resistance"
},
{
"type": "drug",
"drug": "levofloxacin",
"confers": "resistance"
},
{
"type": "drug",
"drug": "moxifloxacin",
"confers": "resistance"
},
{
"type": "drug",
"drug": "ofloxacin",
"confers": "resistance"
}
],
"genome_positions": [
6749,
6750,
6751
]
}
},
"Rv2043c": {
"p.Thr100Ile": {
"annotations": [
{
"type": "drug",
"drug": "pyrazinamide",
"literature": "10.1128/JCM.01214-17",
"confers": "resistance"
}
],
"genome_positions": [
2288942,
2288943,
2288944
]
},
"p.Thr160Ala": {
"annotations": [
{
"type": "drug",
"drug": "pyrazinamide",
"literature": "10.1128/JCM.01214-17",
"confers": "resistance"
}
],
"genome_positions": [
2288762,
2288763,
2288764
]
},
"c.101_102insT": {
"annotations": [
{
"type": "drug",
"drug": "pyrazinamide",
"confers": "resistance"
}
],
"genome_positions": [
2289140,
2289141
]
}
}
}';
-- test if it is a legit JSON
SELECT ISJSON(@json) AS Result;
SELECT genes.[Key] AS gene
, mutations.[Key] AS mutation
, annotations.*
, JSON_VALUE(mutations.value, '$.genome_positions[0]') as [gen_pos1]
, JSON_VALUE(mutations.value, '$.genome_positions[1]') as [gen_pos2]
, JSON_VALUE(mutations.value, '$.genome_positions[2]') as [gen_pos3]
FROM OPENJSON (@json) AS genes
CROSS APPLY OPENJSON(genes.value) AS mutations
CROSS APPLY OPENJSON(mutations.value, '$.annotations')
WITH
(
[type] VARCHAR(20) '$.type'
, [drug] VARCHAR(20) '$.drug'
, [literature] VARCHAR(200) '$.literature'
, [confers] VARCHAR(20) '$.confers'
) AS annotations
Output
--------- --------------- ------ ------------------ ------------------------------------------- ------------ ---------- ---------- ----------
| gene | mutation | type | drug | literature | confers | gen_pos1 | gen_pos2 | gen_pos3 |
--------- --------------- ------ ------------------ ------------------------------------------- ------------ ---------- ---------- ----------
| Rv0005 | p.Glu540Asp | drug | moxifloxacin | 10.1128/AAC.00825-17;10.1128/JCM.06860-11 | resistance | 6857 | 6858 | 6859 |
| Rv0005 | p.Ala504Thr | drug | ciprofloxacin | NULL | resistance | 6749 | 6750 | 6751 |
| Rv0005 | p.Ala504Thr | drug | fluoroquinolones | NULL | resistance | 6749 | 6750 | 6751 |
| Rv0005 | p.Ala504Thr | drug | levofloxacin | NULL | resistance | 6749 | 6750 | 6751 |
| Rv0005 | p.Ala504Thr | drug | moxifloxacin | NULL | resistance | 6749 | 6750 | 6751 |
| Rv0005 | p.Ala504Thr | drug | ofloxacin | NULL | resistance | 6749 | 6750 | 6751 |
| Rv0005 | p.Ala504Val | drug | ciprofloxacin | NULL | resistance | 6749 | 6750 | 6751 |
| Rv0005 | p.Ala504Val | drug | fluoroquinolones | NULL | resistance | 6749 | 6750 | 6751 |
| Rv0005 | p.Ala504Val | drug | levofloxacin | NULL | resistance | 6749 | 6750 | 6751 |
| Rv0005 | p.Ala504Val | drug | moxifloxacin | NULL | resistance | 6749 | 6750 | 6751 |
| Rv0005 | p.Ala504Val | drug | ofloxacin | NULL | resistance | 6749 | 6750 | 6751 |
| Rv2043c | p.Thr100Ile | drug | pyrazinamide | 10.1128/JCM.01214-17 | resistance | 2288942 | 2288943 | 2288944 |
| Rv2043c | p.Thr160Ala | drug | pyrazinamide | 10.1128/JCM.01214-17 | resistance | 2288762 | 2288763 | 2288764 |
| Rv2043c | c.101_102insT | drug | pyrazinamide | NULL | resistance | 2289140 | 2289141 | NULL |
--------- --------------- ------ ------------------ ------------------------------------------- ------------ ---------- ---------- ----------
CodePudding user response:
Using a temporary table will make it easier to pivot the data from the unfolded json.
DECLARE @txt1 varchar(max) = '{...}' IF OBJECT_ID('tempdb..#tmpJsonUnfolded', 'U') IS NOT NULL DROP TABLE #tmpJsonUnfolded; SELECT lvl1.[key] as gene , lvl2.[key] as mutations , lvl3.[key] as data_class , lvl4.[key] as num , lvl5.[key] as col , case when lvl3.[key] = 'genome_positions' then lvl4.[value] when lvl3.[key] = 'annotations' then lvl5.[value] end as [value] --, lvl4.[value] as value4 --, lvl5.[value] as value5 INTO #tmpJsonUnfolded FROM OPENJSON(@txt1) lvl1 CROSS APPLY OPENJSON(lvl1.value) lvl2 CROSS APPLY OPENJSON(lvl2.value) lvl3 CROSS APPLY OPENJSON(lvl3.value) lvl4 OUTER APPLY ( SELECT * FROM OPENJSON(lvl4.value) WHERE lvl3.[key] = 'annotations' ) lvl5;
select gene , mutations , [type] as annotation_type , [num] as annotation_num , [drug] as annotation_drug , [literature] as annotation_literature , [confers] as annotation_confers , [genome_positions] from ( select gene , mutations , num , [col] , [value] from #tmpJsonUnfolded where data_class = 'annotations' union all select gene , mutations , 0 , data_class as [col] , string_agg([value], ', ') as [value] from #tmpJsonUnfolded where data_class = 'genome_positions' group by gene, mutations, data_class ) src pivot ( max([value]) for [col] in ([type], [drug], [literature], [confers], [genome_positions]) ) pvt
gene | mutations | annotation_type | annotation_num | annotation_drug | annotation_literature | annotation_confers | genome_positions :------ | :------------ | :-------------- | -------------: | :--------------- | :---------------------------------------- | :----------------- | :------------------------ Rv0005 | p.Ala504Thr | drug | 0 | ciprofloxacin | null | resistance | 6749, 6750, 6751 Rv0005 | p.Ala504Thr | drug | 1 | fluoroquinolones | null | resistance | null Rv0005 | p.Ala504Thr | drug | 2 | levofloxacin | null | resistance | null Rv0005 | p.Ala504Thr | drug | 3 | moxifloxacin | null | resistance | null Rv0005 | p.Ala504Thr | drug | 4 | ofloxacin | null | resistance | null Rv0005 | p.Ala504Val | drug | 0 | ciprofloxacin | null | resistance | 6749, 6750, 6751 Rv0005 | p.Ala504Val | drug | 1 | fluoroquinolones | null | resistance | null Rv0005 | p.Ala504Val | drug | 2 | levofloxacin | null | resistance | null Rv0005 | p.Ala504Val | drug | 3 | moxifloxacin | null | resistance | null Rv0005 | p.Ala504Val | drug | 4 | ofloxacin | null | resistance | null Rv0005 | p.Glu540Asp | drug | 0 | moxifloxacin | 10.1128/AAC.00825-17;10.1128/JCM.06860-11 | resistance | 6857, 6858, 6859 Rv2043c | c.101_102insT | drug | 0 | pyrazinamide | null | resistance | 2289140, 2289141 Rv2043c | p.Thr100Ile | drug | 0 | pyrazinamide | 10.1128/JCM.01214-17 | resistance | 2288942, 2288943, 2288944 Rv2043c | p.Thr160Ala | drug | 0 | pyrazinamide | 10.1128/JCM.01214-17 | resistance | 2288762, 2288763, 2288764
Demo on db<>fiddle here