I'm looking to set up a population dynamics model where each parameter value corresponds to the temperature of that day. e.g.
Simple model
library(deSolve)
set.seed(1)
pars <- c(alpha = 1, beta = 0.2, delta = 0.5, gamma = 0.2)
lv_model <- function(pars, times = seq(0, 50, by = 1)) {
# initial state
state <- c(x = 1, y = 2)
# derivative
deriv <- function(t, state, pars) {
with(as.list(c(state, pars)), {
d_x <- alpha * x - beta * x * y
d_y <- delta * beta * x * y - gamma * y
return(list(c(x = d_x, y = d_y)))
})
}
# solve
ode(y = state, times = times, func = deriv, parms = pars)
}
lv_results <- lv_model(pars = pars, times = seq(0, 50, by = 1))
I now want to use a sequence of daily temperatures
DailyTemperature<-floor(runif(50,0,40))
and make the parameter values functions of temperatures
TraitTemperature<-seq(1,40,1)
#trait responses to temperature
alpha<- abs(rnorm(40,mean = 0.5,sd=1))
beta<- abs(rnorm(40,mean = 0.2,sd=0.5))
delta<-abs(rnorm(40,mean=1,sd=2))
gamma<- seq(0.025,1,0.025)
parameters<-as.data.frame(cbind(TraitTemperature,alpha,beta,delta,gamma))
So that for each time step iterated over, it looks at the daily temperature and then finds the corresponding temperature values in the parameter data frame.
Looking back through the archives i've seen if/else statements used when wanting to alter single parameters at particular time steps and the use of forcing functions but I don't think they apply here.
I hope this makes sense, I'm interesting in ideas on how to make it work. So far i've also attempted using a for loop to iterate through the daily temperature list and then the match function to identify values but this didn't tap into the daily time steps.
CodePudding user response:
Here a possible approach using DailyTemperature as forcing and then the parameters data frame as lookup table. It does not need match here as long as the temperatures are integers and the temperatures in the data frame correspond to the row index of the data frame.
I want to add that, in principle, discontinuous forcings make the model slow, because an ODE is a continuous by definition. Fortunately, as the solvers are quite robust, it should for practical applications:
library(deSolve)
set.seed(1)
deriv <- function(t, state, pars) {
pars <- parameters[DailyTemperature[floor(t 1)], 2:5]
#print(pars)
with(as.list(c(state, pars)), {
d_x <- alpha * x - beta * x * y
d_y <- delta * beta * x * y - gamma * y
list(c(x = d_x, y = d_y), alpha=alpha, beta=beta, gamma=gamma, delta=delta)
})
}
state <- c(x = 1, y = 2)
times = seq(0, 50, by = 1)
# pars <- c(alpha = 1, beta = 0.2, delta = 0.5, gamma = 0.2)
parameters <- data.frame(
TraitTemperature = seq(1,40,1),
alpha = abs(rnorm(40,mean = 0.5,sd=1)),
beta = abs(rnorm(40,mean = 0.2,sd=0.5)),
delta = abs(rnorm(40,mean=1,sd=2)),
gamma = seq(0.025,1,0.025)
)
DailyTemperature <- floor(runif(51, 0, 40)) # one more because start zero
out <- ode(y = state, times = times, func = deriv, parms = pars)
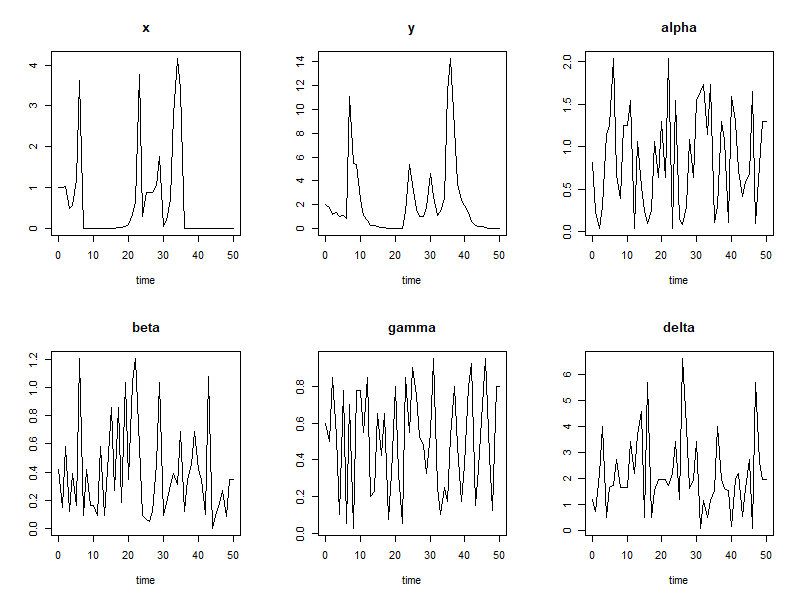
plot(out)
Parameters as a list
In the example above, the pars variable passed from ode is just overwritten with pars derived from the global variables parameters and DailyTemperature. This works, but one may also consider to pass both as a list to the deriv function.
deriv <- function(t, state, p) {
parameters <- p[[1]]
DailyTemperature <- p[[2]]
parms <- parameters[DailyTemperature[floor(t 1)], 2:5]
# ...
}
and then:
out <- ode(y = state, times = times, func = deriv,
parms = list(parameters, DailyTemperature))
CodePudding user response:
This seems to work for indexing using the current reproducible code:
set.seed(1)
deriv <- function(t, state, pars) {
pars<- parameters[match(parameters$TraitTemperature[parameters[2:5]],DailyTemperature),]
#print(pars)
with(as.list(c(state, pars)), {
d_x <- alpha * x - beta * x * y
d_y <- delta * beta * x * y - gamma * y
list(c(x = d_x, y = d_y), alpha=alpha, beta=beta, gamma=gamma, delta=delta)
})
}
state <- c(x = 1, y = 2)
times = seq(0, 50, by = 1)
# pars <- c(alpha = 1, beta = 0.2, delta = 0.5, gamma = 0.2)
parameters <- data.frame(
TraitTemperature = seq(1,40,1),
alpha = abs(rnorm(40,mean = 0.5,sd=1)),
beta = abs(rnorm(40,mean = 0.2,sd=0.5)),
delta = abs(rnorm(40,mean=1,sd=2)),
gamma = seq(0.025,1,0.025)
)
DailyTemperature <- floor(runif(51, 0, 40)) # one more because start zero
out <- ode(y = state, times = times, func = deriv, parms = pars)
plot(out)
but if I increase the resolution of the values e.g.
# Parameter datasets
parameters <- data.frame(
TraitTemperature = seq(0.1,40,0.1),
alpha = abs(rnorm(400,mean = 0.5,sd=1)),
beta = abs(rnorm(400,mean = 0.2,sd=0.5)),
delta = abs(rnorm(400,mean=1,sd=2)),
gamma = seq(0.0025,1,0.0025)
)
# random daily temperature dataset
DailyTemperature <- round(runif(51, 0, 40),1) # one more because start zero
Then I get NA's at certain time steps
CodePudding user response:
The OP extended her/his question, so it may be the preferred way to start a new thread, but in order to give quick feedback, I try give another answer.
Several methods exist for interpolation or indexing in 2D (time and temperature). My preferred way would be to create a 2D model and then tu use a 2D interpolation method. This works best if the parameters surface would be smooth and not just random as in the given example. However, to make it simple, one can also use rounding and table lookup. If values are not integers, exact comparison does often not work due to rounding effects and limited precision (IEEE number formats), so instead of match, one can use which.min:
DailyTemperature <- round(runif(51, 0, 40), 1)
TraitTemperature <- seq(0, 40, by=0.1)
N <- length(TraitTemperature)
parameters <- data.frame(
TraitTemperature = TraitTemperature,
alpha = abs(rnorm(N, mean = 0.5, sd=1)),
beta = abs(rnorm(N, mean = 0.2, sd=0.5)),
delta = abs(rnorm(N, mean=1,sd=2)),
gamma = seq(0.025, 1, length.out=N)
)
t <- 17
actualTemp <- DailyTemperature[floor(t 1)]
actualTemp
pars <- parameters[which.min(abs(actualTemp - parameters$TraitTemperature)), 1:5]
head(pars)
