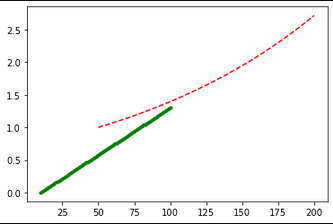
Suppose you have the following data:
x1_Length = 100
x1 = np.linspace(10,100,num=x1_Length)
y1 = np.linspace(0, 1.3, num=x1_Length)
x2_Length = 200
x2 = np.linspace(50, 200, num=x2_Length)
y2 = np.exp(np.linspace(0, 1, num=x2_Length))
If you wanted to plot this, it would be straightforward:
fig, ax = plt.subplots()
ax.plot(x1,y1, ".g")
ax.plot(x2,y2, "--r")
Which gives an expected result, EVEN THOUGH THE ARRAYS THAT WERE PLOTTED ARE OF DIFFERENT LENGTHS AND DO NOT SHARE THE x-data.
NOW, suppose you wanted to plot this same data in an intensity plot. This causes some problems:
it is my understanding that an intensity plot (for example
imshow) plots 2D arrays and as such have no direct concept of an x-axis scale or y-axis scale. Data is presented based on its index in the array. Here, this would be a problem because the first elements of the two arrays DO NOT correspond to the same x-value.all the rows/columns need to be of the same length in a 2D array. This is the minor of the two problems, as i could always figure out the longest dataset and pad all shorter ones with, for example, NaNs, like so:
dataset = [x1_data, x2_data] max_Length = np.max([array[0,:].shape[0] for array in dataset ]) data_array = np.nan * np.ones((2*len(dataset), max_Length)) for i, spectrum_array2D in enumerate(dataset): length = spectrum_array2D[0,:].shape[0] firstRowOfInsertion = 2*i data_array[[firstRowOfInsertion, firstRowOfInsertion 1], :length] = spectrum_array2D
I don't see any technological limitation why one should not be able to plot several arrays of different data into an intensity plot much like plt.plot()can. Ideally, what I need is to tell the intensity plot that it can expect, in this example, 2 horizontal slices of data, and then supply the x-data (plotted againt the x-axis), the y-data (plotted against the colorbar) for each index of the slice into which this plot should go. Do you know of a way to achieve this, or any package capable of this?
CodePudding user response:
One option with matplotlib is pcolormesh: 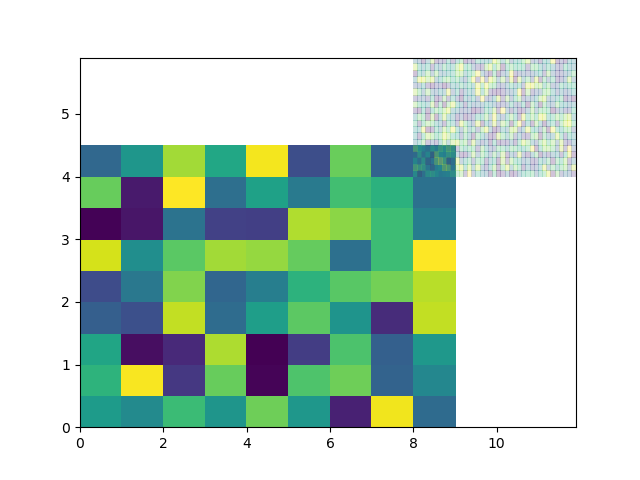
CodePudding user response:
As far as I understand, you are aware, that the two datasets can not intersect each other. That's the first important condition. If to values would occupy the same pixel in the meshgrid, which one would be chosen? But even if the data does not intersect, there could be a problem when both axis have different sampling rates/distances. Before you could plot both of them in the same image or even mesh, you would have to resample/interpolate both arrays on an equal grid for both. This get's really tricky quite quickly and is prone to errors. If I were to try this, I would start by creating to seperate color maps, with the same colorcode, but two indipendent meshs. The next step would be to just lay one of figures on top of the other and shift it so that the axis line up; basically an inset. I suppose there is some best practice for that in matplotlib.