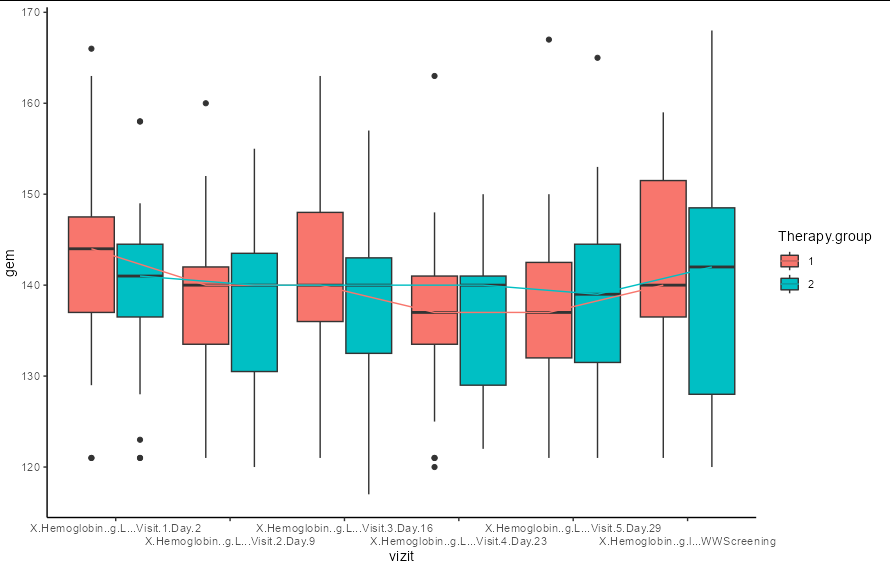
I need create plot using ggplot2
Here my code with reproducible example
library(tidyverse)
gemoglabin=structure(list(Therapy.group = c(1L, 2L, 1L, 2L, 1L, 2L, 2L,
2L, 1L, 1L, 2L, 1L, 2L, 2L, 1L, 1L, 1L, 2L, 2L, 1L, 2L, 1L, 2L,
1L, 2L, 2L, 2L, 1L, 1L, 2L, 1L, 2L, 2L, 1L, 1L, 2L, 1L, 1L),
X.Hemoglobin..g.l...WWScreening = c(139L, 168L, 152L, 122L,
142L, 150L, 120L, 157L, 136L, 128L, 157L, 154L, 142L, 124L,
138L, 138L, 151L, 148L, 148L, 142L, 149L, 157L, 137L, 135L,
121L, 148L, 121L, 159L, 133L, 132L, 155L, 133L, 136L, 121L,
140L, 142L, 137L, 147L), X.Hemoglobin..g.L...Visit.1.Day.2 = c(166L,
141L, 144L, 121L, 141L, 145L, 121L, 158L, 135L, 121L, 158L,
152L, 144L, 128L, 129L, 146L, 142L, 141L, 139L, 139L, 140L,
159L, 143L, 132L, 134L, 149L, 123L, 163L, 145L, 141L, 145L,
142L, 146L, 121L, 143L, 139L, 145L, 149L), X.Hemoglobin..g.L...Visit.2.Day.9 = c(140L,
155L, 142L, 122L, 138L, 143L, 126L, 151L, 129L, 124L, 155L,
140L, 140L, 122L, 132L, 136L, 135L, 140L, 141L, 142L, 136L,
145L, 138L, 126L, 128L, 144L, 120L, 160L, 143L, 133L, 141L,
142L, 147L, 121L, 138L, 139L, 142L, 152L), X.Hemoglobin..g.L...Visit.3.Day.16 = c(150L,
157L, 153L, 117L, 140L, 143L, 122L, 155L, 125L, 121L, 156L,
144L, 137L, 122L, 136L, 136L, 140L, 140L, 143L, 143L, 140L,
143L, 141L, 123L, 129L, 150L, 121L, 163L, 149L, 143L, 136L,
136L, 143L, 123L, 147L, 137L, 139L, 152L), X.Hemoglobin..g.L...Visit.4.Day.23 = c(140L,
150L, 144L, 122L, 139L, 140L, 124L, 149L, 121L, 125L, 148L,
141L, 132L, 126L, 133L, 136L, 137L, 141L, 141L, 137L, 141L,
145L, 140L, 120L, 124L, 143L, 122L, 163L, 139L, 132L, 141L,
141L, 138L, 121L, 134L, 134L, 137L, 148L), X.Hemoglobin..g.L...Visit.5.Day.29 = c(132L,
165L, 141L, 122L, 143L, 140L, 131L, 153L, 123L, 121L, 150L,
150L, 139L, 132L, 132L, 139L, 136L, 134L, 145L, 137L, 144L,
147L, 134L, 124L, 128L, 148L, 121L, 167L, 142L, 127L, 140L,
141L, 144L, 126L, 132L, 134L, 136L, 148L)), class = "data.frame", row.names = c(NA,
-38L))
datadim.lng <- gemoglabin %>% pivot_longer(-Therapy.group,names_to="vizit", values_to = "gem")
datadim.lng$Therapy.group <- as.factor(datadim.lng$Therapy.group)
ggplot(datadim.lng, aes(x = vizit, y = gem, fill = Therapy.group))
geom_boxplot(position = position_dodge(width = 0.85), width = 0.8)
theme_classic()
stat_summary(
fun = median,
geom = 'line',
aes(group = Therapy.group, color = Therapy.group),
position = position_dodge(width = 0.85)
)
as the result i get such screen
as we can see here the signatures on the vizit axis are mixed up and i can’t really see which vizit number.
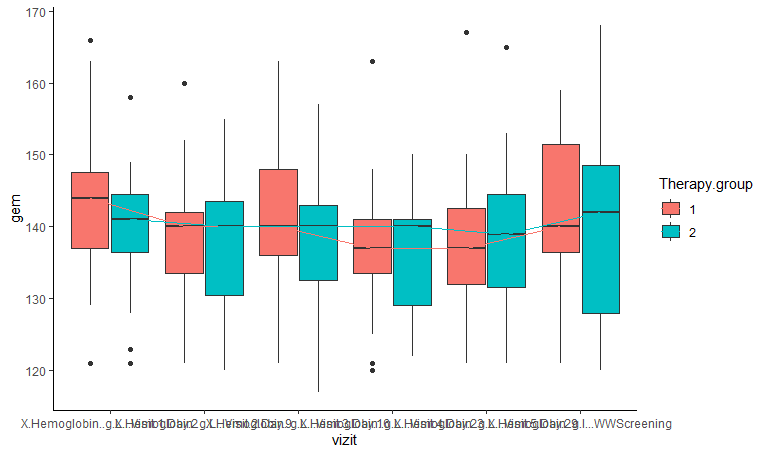
How to make the signatures not mixed and visible? I must have space between visits in plot
only not in such small print otherwise it is hard to see.
Another problem is wrong order of signatures. WWscreening as we can see the first column in dataset ,but in the plot it is last in order
Can you please help me fix space between signatures and correct order of column in the plot. Thank you
CodePudding user response:
You can use stringr::str_wrap within scale_x_discrete to wrap your labels, and fct_inorder to convert your column to ordered factor by the order in which they appear.
datadim.lng %>%
mutate(vizit = fct_inorder(vizit)) %>% # convert to factor in order of appearance
ggplot(aes(x = vizit, y = gem, fill = Therapy.group))
geom_boxplot(position = position_dodge(width = 0.85), width = 0.8)
theme_classic()
stat_summary(
fun = median,
geom = 'line',
aes(group = Therapy.group, color = Therapy.group),
position = position_dodge(width = 0.85)
)
scale_x_discrete(labels = function(x) str_wrap(gsub("\\. ", " ", x), width = 5))
the labels argument can be provided with either the labels to use or a function that will be applied to all labels. In the above code a function is created that firsts convert the dots to spaces within your column and then uses str_wrap for wrapping the words into multiple lines. (str_wrap won't work if there are no breaks between words)
CodePudding user response:
I think there are two answers here. One is the solution to the question you asked, which is that you can prevent clashes using guide_axis(n.dodge = 2)
ggplot(datadim.lng, aes(x = vizit, y = gem, fill = Therapy.group))
geom_boxplot(position = position_dodge(width = 0.85), width = 0.8)
theme_classic()
stat_summary(
fun = median,
geom = 'line',
aes(group = Therapy.group, color = Therapy.group),
position = position_dodge(width = 0.85)
)
guides(x = guide_axis(n.dodge = 2))
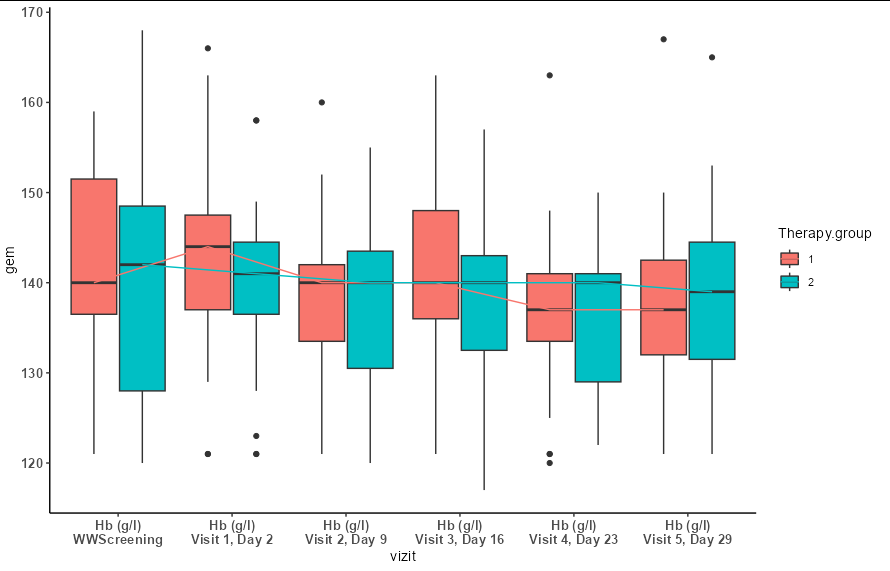
But I think a better answer is that you should rename the levels you are plotting. This will make a plot that is easier for the viewer to parse and understand.
datadim.lng$vizit <- gsub("X\\.Hemoglobin\\.\\.g\\.(l|L)", "Hb (g/l)",
datadim.lng$vizit)
datadim.lng$vizit <- gsub("\\.{3}", "\n", datadim.lng$vizit)
datadim.lng$vizit <- gsub("\\.", " ", datadim.lng$vizit)
datadim.lng$vizit <- gsub(" D", ", D", datadim.lng$vizit)
datadim.lng$vizit <- factor(datadim.lng$vizit,
levels(factor(datadim.lng$vizit))[c(6, 1:5)])
ggplot(datadim.lng, aes(x = vizit, y = gem, fill = Therapy.group))
geom_boxplot(position = position_dodge(width = 0.85), width = 0.8)
theme_classic()
stat_summary(
fun = median,
geom = 'line',
aes(group = Therapy.group, color = Therapy.group),
position = position_dodge(width = 0.85)
)
theme(axis.text = element_text(size = 10, face = 2))