I want to perform the following task using fastfood dataset from openintro package in R.
a) Create a regression predicting whether or not a restaurant is McDonalds or Subway based on calories, sodium, and protein. (McDonalds should be 1, Subway 0).
Save the coefficients to Q2.
b) use data from only restaurants with between 50 and 60 items in the data set. Predict total fat from cholesterol, total carbs, vitamin a, and restaurant. Remove any nonsignificant predictors and run again.
Assign the strongest standardized regression coefficient to Q5.
Here's my code.
library(tidyverse)
library(openintro)
library(lm.beta)
fastfood <- openintro::fastfood
head(fastfood)
#Solving for part (a)
fit_1 <- lm(I(restaurant %in% c("Subway", "Mcdonalds")) ~ calories sodium protein, data = fastfood)
Q2 <- round(summary(fit_1)$coefficients,2)
#Solving for part (b)
newdata <- fastfood[ which(fastfood$item>=50 & fastfood$item <= 60), ]
df = sort(sample(nrow(newdata), nrow(data)*.7))
newdata_train<-data[df,]
newdata_test<-data[-df,]
fit_5 <- lm(I(total_fat) ~ cholesterol total_carb vit_a restaurant, data = newdata)
prediction_5 <- predict(fit_5, newdata = newdata_test)
Q5 <- lm.beta(fit_5)
But I'm not getting desired results
Here's is desired output
output for part (a):
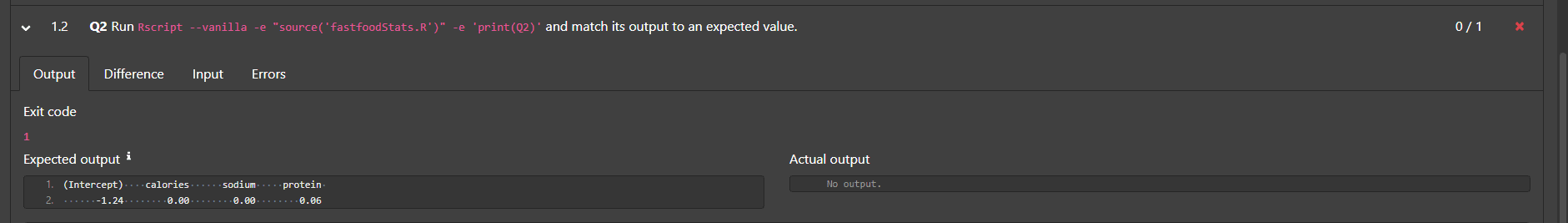
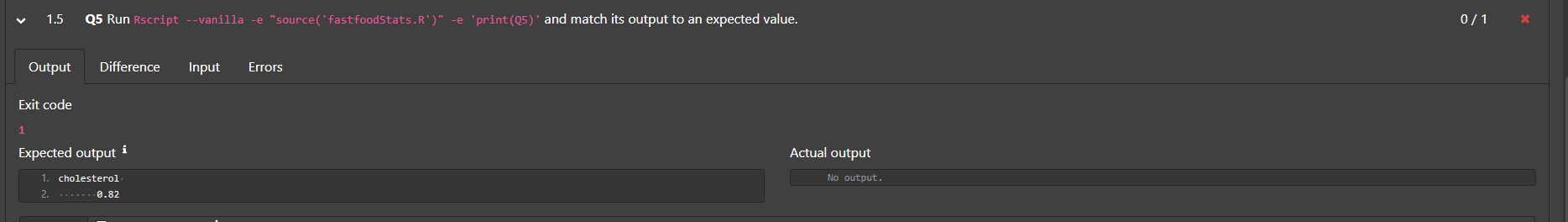
output for part (b):
CodePudding user response:
The first question requires logistic regression rather than linear regression, since the aim is to predict a binary outcome. The most sensible way to do this is, as the question suggests, to remove all the restaurants except McDonald's and Subway, then create a new binary variable to mark which rows are McDonald's and which aren't:
library(dplyr)
fastfood <- openintro::fastfood %>%
filter(restaurant %in% c("Mcdonalds", "Subway")) %>%
mutate(is_mcdonalds = restaurant == "Mcdonalds")
The logistic regression is done like this:
fit_1 <- glm(is_mcdonalds ~ calories sodium protein,
family = "binomial", data = fastfood)
And your coefficients are obtained like this:
Q2 <- round(coef(fit_1), 2)
Q2
#> (Intercept) calories sodium protein
#> -1.24 0.00 0.00 0.06
The second question requires that you filter out any restaurants with more than 60 or fewer than 50 items:
fastfood <- openintro::fastfood %>%
group_by(restaurant) %>%
filter(n() >= 50 & n() <= 60)
We now fit the described regression and examine it to look for non-significant regressors:
fit_2 <- lm(total_fat ~ cholesterol vit_a total_carb restaurant,
data = fastfood)
summary(fit_2)
#>
#> Call:
#> lm(formula = total_fat ~ cholesterol vit_a total_carb restaurant,
#> data = fastfood)
#>
#> Residuals:
#> Min 1Q Median 3Q Max
#> -24.8280 -2.9417 0.9397 5.1450 21.0494
#>
#> Coefficients:
#> Estimate Std. Error t value Pr(>|t|)
#> (Intercept) -1.20102 2.08029 -0.577 0.564751
#> cholesterol 0.26932 0.01129 23.853 < 2e-16 ***
#> vit_a 0.01159 0.01655 0.701 0.484895
#> total_carb 0.16327 0.03317 4.922 2.64e-06 ***
#> restaurantMcdonalds -4.90272 1.94071 -2.526 0.012778 *
#> restaurantSonic 6.43353 1.89014 3.404 0.000894 ***
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
#>
#> Residual standard error: 7.611 on 125 degrees of freedom
#> (34 observations deleted due to missingness)
#> Multiple R-squared: 0.8776, Adjusted R-squared: 0.8727
#> F-statistic: 179.2 on 5 and 125 DF, p-value: < 2.2e-16
We note that vit_a is non-significant and drop it from our model:
fit_3 <- update(fit_2, . ~ . - vit_a)
Now we get the regularized coefficients and round them:
coefs <- round(coef(lm.beta::lm.beta(fit_3)), 2)
and Q5 will be the maximum value of these coefficients:
Q5 <- coefs[which.max(coefs)]
Q5
#> cholesterol
#> 0.82
Created on 2022-02-26 by the reprex package (v2.0.1)
