I want to get the mean of every interval with values above a threshold. Obviously, I could do a loop and just look if the next value is under the threshold etc., but I was hoping that there would be an easier way. Do you have ideas that are similar to something like masking, but include the "interval"-problem?
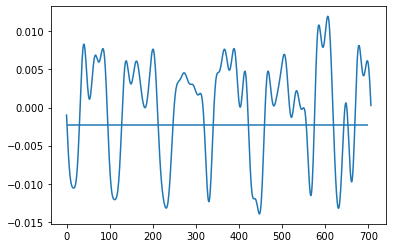
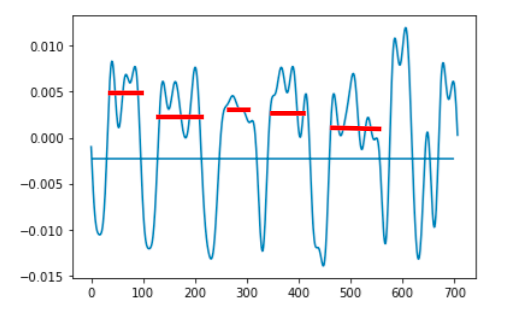
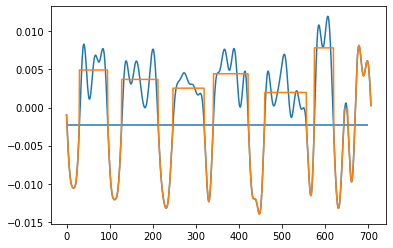
Below are 2 pictures with the original data and what I want to obtain.
Before:
After:
My original idea was looping through my array, but as I want to do this about 10.000 times or more, I guess it's getting very time intensive.
Is there a way to get rid of the for loops?
transformed is a numpy array.
plt.figure()
plt.plot(transformed)
thresh=np.percentile(transformed,30)
plt.hlines(np.percentile(transformed,30),0,700)
transformed_copy=transformed
transformed_mask=[True if x>thresh else False for x in transformed_copy]
mean_arr=[]
for k in range(0,len(transformed)):
if transformed_mask[k]==False:
mean_all=np.mean(transformed_copy[mean_arr])
for el in mean_arr:
transformed_copy[el]=mean_all
mean_arr=[]
if transformed_mask[k]==True:
mean_arr.append(k)
plt.plot(transformed_copy)
Output after loop:
CodePudding user response:
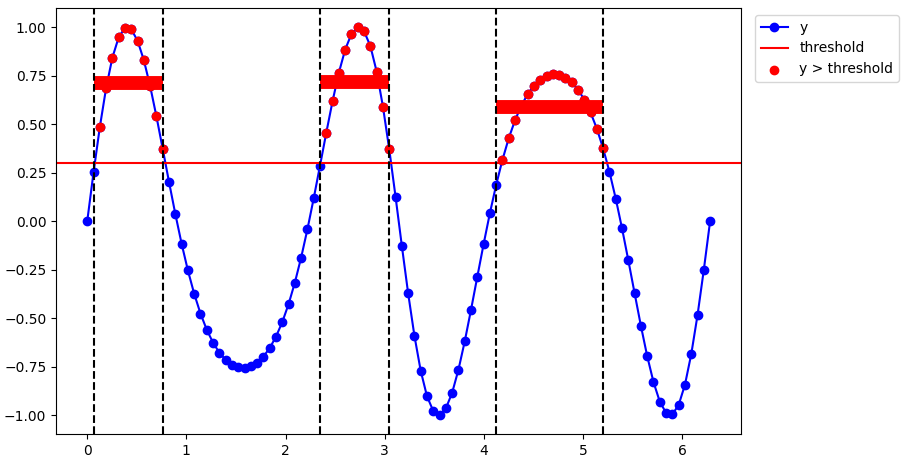
The trick I am using here is to calculate where there are sudden differences in the mask which means we switch from a contiguous section to another. Then we get the indexes of where those sections start and end, and calculate the mean inside of them.
# Imports.
import matplotlib.pyplot as plt
import numpy as np
# Create data.
x = np.linspace(0, 2*np.pi, 100)
y = np.sin(np.sin(x)*4)
threshold = 0.30
mask = y > threshold
# Plot the raw data, threshold, and show where the data is above the threshold.
fig, ax = plt.subplots()
ax.plot(x, y, color="blue", label="y", marker="o", zorder=0)
ax.scatter(x[mask], y[mask], color="red", label="y > threshold", zorder=1)
ax.axhline(threshold, color="red", label="threshold")
ax.legend(loc="upper left", bbox_to_anchor=(1.01, 1))
# Detect the different segments.
diff = np.diff(mask) # Where the mask starts and ends.
jumps = np.where(diff)[0] # Indices of where the mask starts and ends.
for jump in jumps:
ax.axvline(x[jump], linestyle="--", color="black")
# Calculate the mean inside each segment.
for n1, n2 in zip(jumps[:-1:2], jumps[1::2]):
xn = x[n1:n2]
yn = y[n1:n2]
mean_in_section_n = np.mean(yn)
ax.hlines(mean_in_section_n, xn[0], xn[-1], color="red", lw=10)
fig.show()
With a bit more time, we could imagine a function that encases all this logic and has this signature: f(data, mask) -> data1, data2, ... with an element returned for each contiguous section.
def data_where_mask_is_contiguous(data:np.array, mask:np.array) -> list:
sections = []
diff = np.diff(mask) # Where the mask starts and ends.
jumps = np.where(diff)[0] # Indices of where the mask starts and ends.
for n1, n2 in zip(jumps[:-1:2], jumps[1::2]):
sections.append(data[n1:n2])
return sections
With this, you can get the mean in each section very easily:
print([np.mean(yn) for yn in data_where_mask_is_contiguous(y, mask)])
>>> [0.745226, 0.747790, 0.599429]
I just noticed it doesn't work when the mask is all true, so I need to add a default case but you get the idea.