I have a data frame with bacteria families from with all their OTUs (phylum, order, family...).
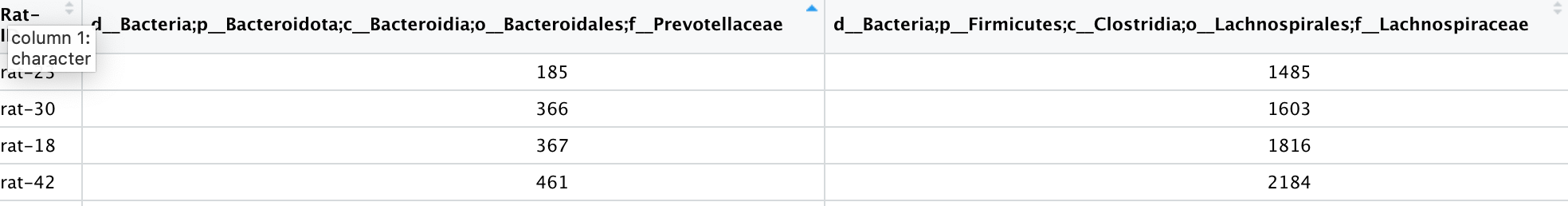
The data frame is large and I would like the name of each column to be only the last part of each string. The one that starts with "f___"
For example
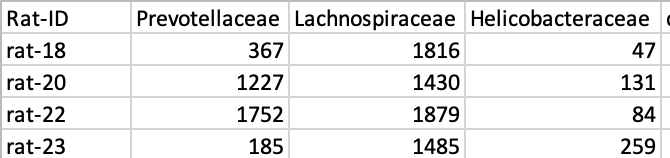
I tried some methods in R (like dplyr::filter or filter(str_detect))and also separating columns in Excel and could not get what I wanted. I don't do it manually because it's too many columns.
Thanks
CodePudding user response:
df being your dataframe, you could use rename_with from package dplyr:
df %>%
rename_with(
## your renaming function (see ?gsub for help on
## replacing with search patterns (regular expressions):
~ gsub('.*;f___(.*)$', '\\1', .x),
## column selection (see ?dplyr::select for handy shortcuts)
cols = everything()
)
the .x in the replacement formula ~ etc. represents the variable argument to the replacement function, in this case the 'old' column name. You'll encounter this 'dot-something' pattern frequently in tidyverse packages.
CodePudding user response:
microbiota <- read_csv("Tablas/nivel5-familia_clean.csv") colnames(microbiota) <- gsub(colnames(microbiota),pattern = '.*f__', replacement = "")
I solve it like this.
