I would like to use ggplot to create multiple density plots that have the same data but that show a specific line highlighted. The table I have is extremely big but has the following columns:
Marker Sample value
X1 4_HVxLF 0.5
X2 4_HVxLF 0.1
...
 .
.
ggplot(Dta, aes(x=value, group=Sample, color = Sample))
geom_density()
xlab("value")
ylab("Density")
theme_classic()
It is not easy to interpret. Then, My aim is to get something like this: 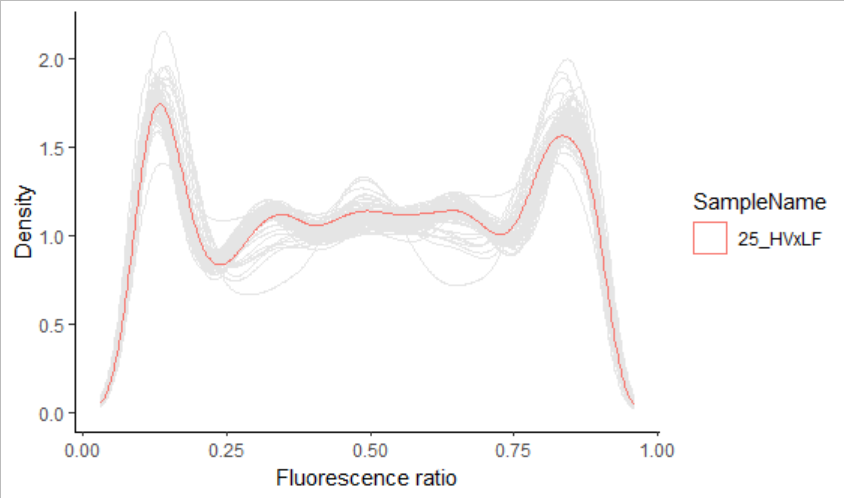 . The code used is:
. The code used is:
ggplot(Dta, aes(x=value, group=Sample, color = Sample))
geom_density()
xlab("value")
ylab("Density")
theme_classic()
gghighlight(SampleName == "25_HVxLF",
unhighlighted_params = list(colour = "grey90"))
So I tried building a loop to get a PDF with 4 plots like the second one per page and for all my samples (105 of them). I cannot used facet_wrap because there are too many of them.
Here is what I tried:
Samples <- unique(Dta$Sample)
pdf('Allplots.pdf', width = 8, height = 11)
par(mfrow=c(4,2))
for (i in Samples){
ggplot(Dta, aes(x=value, group=Sample,
color = Sample))
geom_density()
xlab("value")
ylab("Density")
theme_classic()
gghighlight(Sample == Samples[i],
unhighlighted_params = list(colour = "grey90"))
}
dev.off()
But the PDF that comes out is 4kb and I cannot open it. It says there are no pages. I am not used to work with loops so this is my first attempt ... I don't know what I did wrong (maybe many things), could someone help me figure out ?
Thank you in advance for your kind help !
Diana
CodePudding user response:
Using patchwork and lapply to loop over your samples one approach to achieve your desired result may look like so:
- Use a first loop to create your plots and store them in a list
- Use a second loop to glue the plots together in grids of 2 rows and 2 columns using
patchwork::wrap_plots
Making use of some fake random example data:
library(ggplot2)
library(gghighlight)
library(patchwork)
plot_fun <- function(highlight) {
ggplot(Dta, aes(x=value, group=Sample,
color = Sample))
geom_density()
xlab("value")
ylab("Density")
theme_classic()
gghighlight(Sample == highlight,
unhighlighted_params = list(colour = "grey90"))
}
Samples <- unique(Dta$Sample)
# Make plots
p <- lapply(Samples, plot_fun)
nrow <- 2
ncol <- 2
# Page counter
idx_page <- rep(seq(ceiling(length(p) / nrow / ncol)), each = nrow * ncol, length.out = length(p))
# Export plots as grids of 2 x 2
pdf('Allplots.pdf', width = 8, height = 11)
lapply(unique(idx_page), function(x) {
wrap_plots(p[idx_page == x], ncol = ncol, nrow = nrow)
})
#> [[1]]
#>
#> [[2]]
#>
#> [[3]]
#>
#> [[4]]
#>
#> [[5]]
dev.off()
#> quartz_off_screen
#> 2
DATA
# Random example data
set.seed(123)
n <- 20
Dta <- data.frame(
Sample = rep(LETTERS[seq(n)], each = 100),
value = rnorm(100 * n)
)
