This is a follow up question to 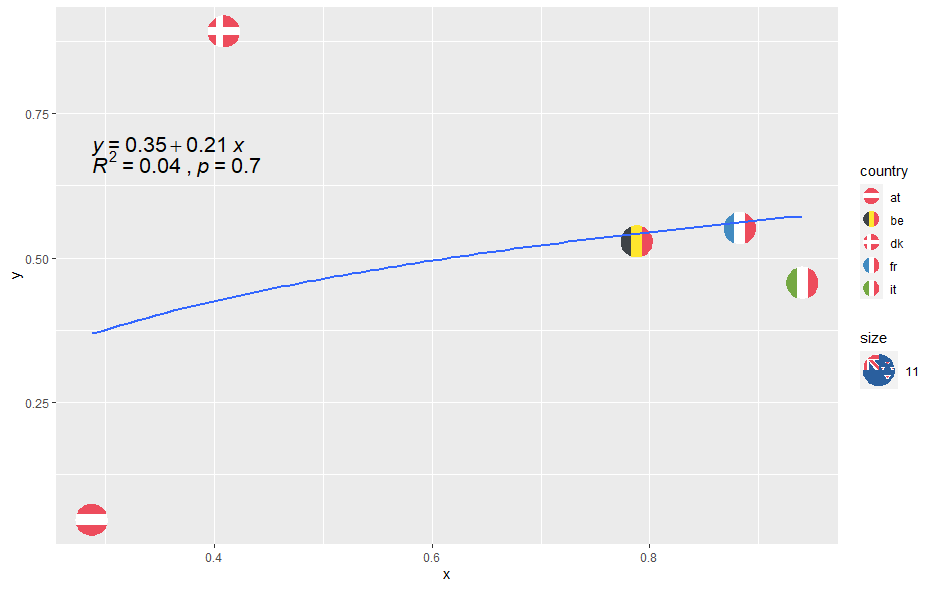
The problem is when I want to print out the regression equation, the R2 and the p-value with the help of stat_regline_equation and stat_cor, I get values for a linear model and not the log-linear model I want to use.
How can I solve this?
library(ggplot2)
library(ggflags)
library(ggpubr)
library(SciViews)
set.seed(123)
Data <- data.frame(
country = c("at", "be", "dk", "fr", "it"),
x = runif(5),
y = runif(5)
)
ggplot(Data, aes(x = x, y = y, country = country, size = 11))
geom_flag()
scale_country()
scale_size(range = c(10, 10))
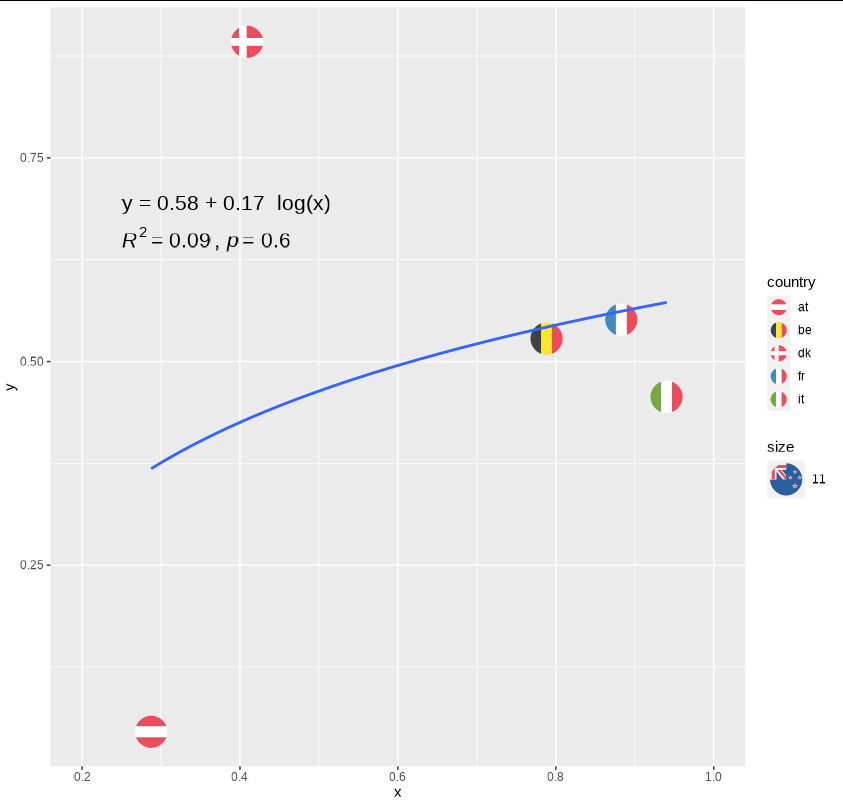
geom_smooth(aes(group = 1), method = "lm", , formula = y ~ log(x), se = FALSE, size = 1)
stat_regline_equation(label.y = 0.695,
aes(group = 1, label = ..eq.label..), size = 5.5)
stat_cor(aes(group = 1,
label =paste(..rr.label.., ..p.label.., sep = "~`,`~")),
label.y = 0.685, size = 5.5, digits= 1)
edit: I have also tried to use ln(x) instead of log(x) but I do not get any results when printing out the coefficient from that either.
CodePudding user response:
There are four things you need to do:
- Provide your regression formula to the
formulaargument ofstat_regline_equation - Use
subto change "x" to "log(x)" ineq.label - Change the x aesthetic of
stat_cortolog(x) - Fix the x limits inside
coord_cartesianto compensate
ggplot(Data, aes(x = x, y = y, country = country, size = 11))
geom_flag()
scale_country()
scale_size(range = c(10, 10))
geom_smooth(aes(group = 1), method = "lm", , formula = y ~ log(x),
se = FALSE, size = 1)
stat_regline_equation(label.y = 0.695, label.x = 0.25,
aes(group = 1, label = sub("x", "log(x)", ..eq.label..)),
size = 5.5,
formula = y ~ log(x),
check_overlap = TRUE, output.type = "latex")
stat_cor(aes(group = 1, x = log(x),
label =paste(..rr.label.., ..p.label.., sep = "~`,`~")),
label.x = 0.25,
label.y = 0.65, size = 5.5, digits= 1, check_overlap = TRUE)
coord_cartesian(xlim = c(0.2, 1))