Please I need help partitioning a symmetric or any matrix in R, say my variables are grouped say the default data in R called longley,
data()
L.df<-longley
cor(L.df)
I need a code which will help me partition this correlation matrix as
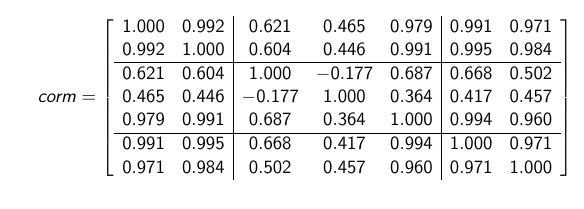
So that I can be able to take the average of it partition allowing me to assume equal correlation as the interpolation of each group!
So I can have something like
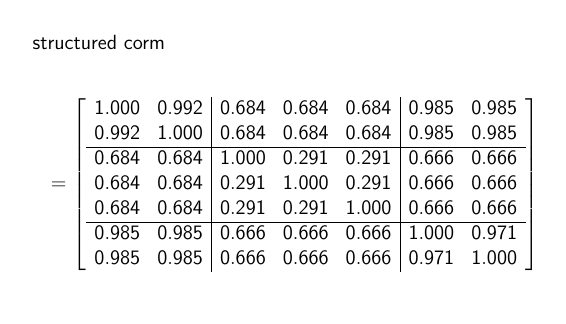
As my structured correlation matrix.
PS. I obtained the structured manually
Would love to be able to partition it at any Column or row of choice.
Note: The partition assumes variables 1&2 are in group 1, Variables 3,4 & 5 are in group 2 And variables 6&7 are in group 3
CodePudding user response:
You can get the correlation matrix, set the diagonal to NA, iterate through the groups and rewrite their entries with their NA-omitted means, then write 1 into the diagonal:
partition <- function(m, groups = list(1:2, 3:5, 6:7)) {
crm <- cor(m)
diag(crm) <- NA
for(i in groups) {
for(j in groups) {
crm[i, j] <- mean(crm[i, j], na.rm = TRUE)
}
}
diag(crm) <- 1
crm <- round(crm, 3)
dimnames(crm) <- NULL
crm
}
partition(longley)
#> [,1] [,2] [,3] [,4] [,5] [,6] [,7]
#> [1,] 1.000 0.992 0.684 0.684 0.684 0.985 0.985
#> [2,] 0.992 1.000 0.684 0.684 0.684 0.985 0.985
#> [3,] 0.684 0.684 1.000 0.291 0.291 0.667 0.667
#> [4,] 0.684 0.684 0.291 1.000 0.291 0.667 0.667
#> [5,] 0.684 0.684 0.291 0.291 1.000 0.667 0.667
#> [6,] 0.985 0.985 0.667 0.667 0.667 1.000 0.971
#> [7,] 0.985 0.985 0.667 0.667 0.667 0.971 1.000
To change the groups, you need to supply them as a list of column indices. For example, if you wanted two groups with columns 1:3 and 4:7, you could do:
partition(longley, list(1:3, 4:7))
#> [,1] [,2] [,3] [,4] [,5] [,6] [,7]
#> [1,] 1.000 0.739 0.739 0.709 0.709 0.709 0.709
#> [2,] 0.739 1.000 0.739 0.709 0.709 0.709 0.709
#> [3,] 0.739 0.739 1.000 0.709 0.709 0.709 0.709
#> [4,] 0.709 0.709 0.709 1.000 0.694 0.694 0.694
#> [5,] 0.709 0.709 0.709 0.694 1.000 0.694 0.694
#> [6,] 0.709 0.709 0.709 0.694 0.694 1.000 0.694
#> [7,] 0.709 0.709 0.709 0.694 0.694 0.694 1.000
Created on 2022-04-11 by the reprex package (v2.0.1)
CodePudding user response:
Define a grouping vector g and from that create a grouping matrix gm and use that with ave.
g <- c(1, 1, 2, 2, 2, 3, 3)
gm <- outer(g, g, paste)
diag(gm) <- ""
ave(cor(longley), gm)
