ggrepel provides an excellent series of functions for annotating ggplot2 graphs and 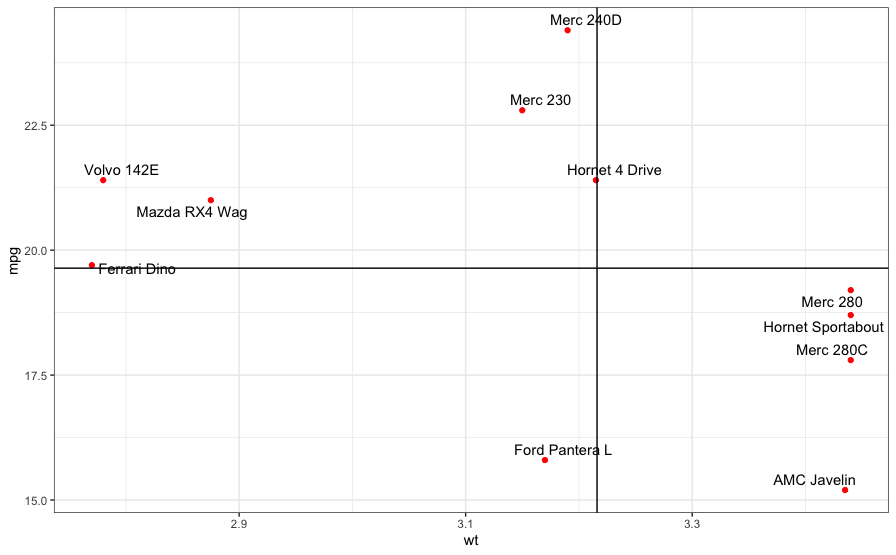
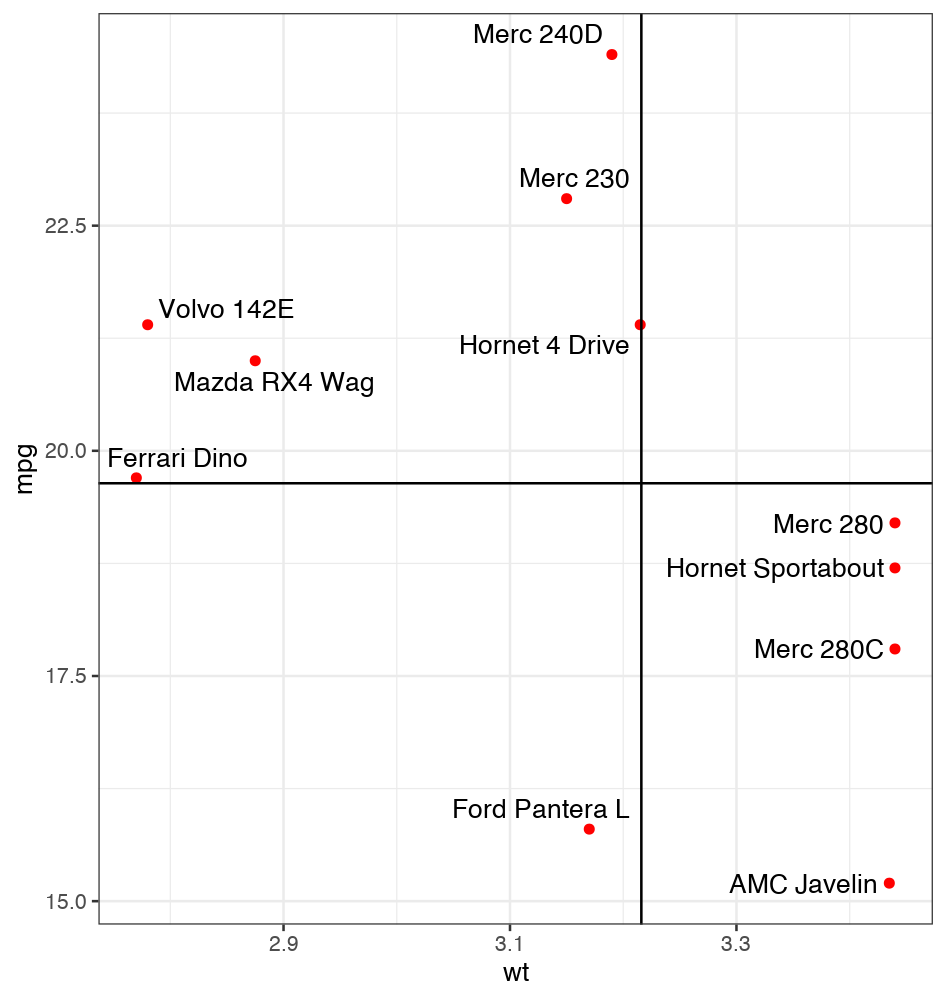
Note how the lines overlap the text of the labels and obscure it (is that "Horret 4 Drive" or "Hornet 4 Drive"?)
Jiggling the points about a bit post facto you can make a far nicer fit – I have simply shifted some of the labels a tiny bit to get them off the line.
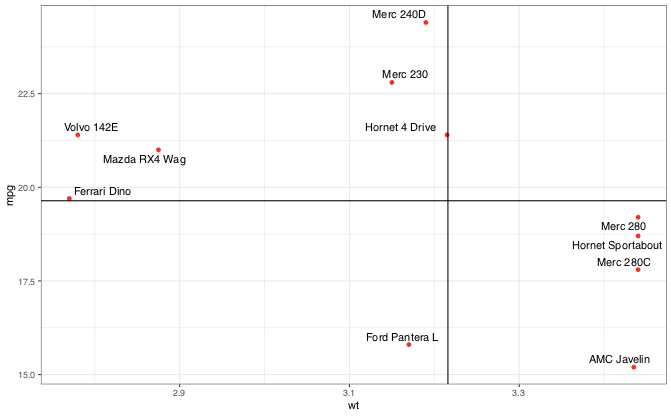
Is it possible to get ggrepel to do this automatically? I know the example given isn't totally stable (other seeds give acceptable results) but for complex plots with a large number of points it definitely is a problem.
Edit: If you're curious, a far less "minimum" working example would be the below (taken from bioconductor):
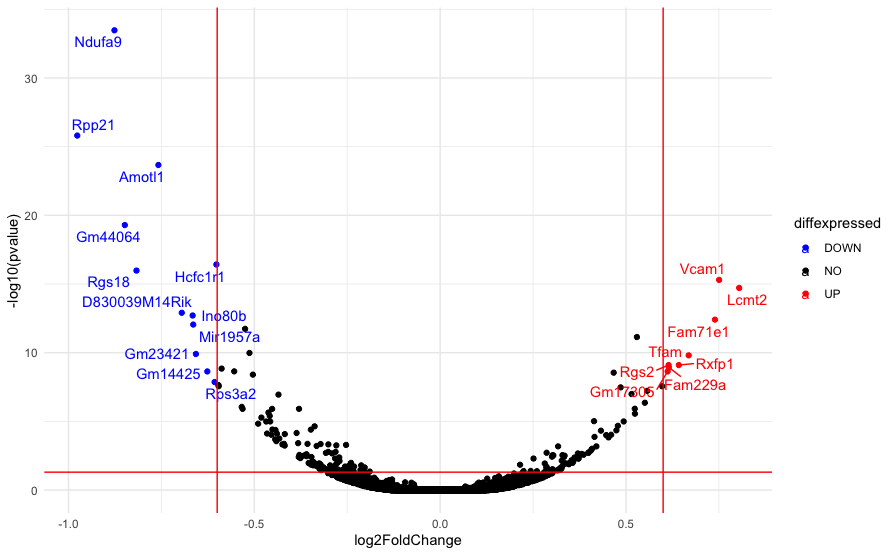
download.file("https://raw.githubusercontent.com/biocorecrg/CRG_RIntroduction/master/de_df_for_volcano.rds", "de_df_for_volcano.rds", method="curl")
tmp <- readRDS("de_df_for_volcano.rds")
de <- tmp[complete.cases(tmp), ]
de$diffexpressed <- "NO"
# if log2Foldchange > 0.6 and pvalue < 0.05, set as "UP"
de$diffexpressed[de$log2FoldChange > 0.6 & de$pvalue < 0.05] <- "UP"
# if log2Foldchange < -0.6 and pvalue < 0.05, set as "DOWN"
de$diffexpressed[de$log2FoldChange < -0.6 & de$pvalue < 0.05] <- "DOWN"
# Create a new column "delabel" to de, that will contain the name of genes differentially expressed (NA in case they are not)
de$delabel <- NA
de$delabel[de$diffexpressed != "NO"] <- de$gene_symbol[de$diffexpressed != "NO"]
#Actually do plot
ggplot(data=de, aes(x=log2FoldChange, y=-log10(pvalue), col=diffexpressed, label=delabel))
geom_point()
theme_minimal()
geom_text_repel()
scale_color_manual(values=c("blue", "black", "red"))
geom_vline(xintercept=c(-0.6, 0.6), col="red")
geom_hline(yintercept=-log10(0.05), col="red")
This produces the below, where the text-overlapping-lines problem is quite obvious:
CodePudding user response:
I don't think there's a built-in way to do this.
A non-elegant hack off the top of my head is to add invisible points along the intercept lines which the labels will then repel away from.
dat <- subset(mtcars, wt > 2.75 & wt < 3.45)
dat$car <- rownames(dat)
xintercept = 3.216
yintercept = 19.64
dat %>%
mutate(alpha = 1) %>%
bind_rows(.,
tibble(wt = seq(from = min(.$wt), to = max(.$wt), length.out = 20), mpg = yintercept, car = '', alpha = 0),
tibble(wt = xintercept, mpg = seq(from = min(.$mpg), to = max(.$mpg), length.out = 20), car = '', alpha = 0)
) %>%
ggplot(aes(wt, mpg, label = car, alpha = alpha))
geom_point(color = "red")
geom_text_repel(seed = 1) #Seed for reproducibility
geom_vline(xintercept = xintercept)
geom_hline(yintercept = yintercept) theme_bw()
scale_alpha_identity()
CodePudding user response:
One (admittedly unorthodox) solution would be to plot "invisible" text along the intercept lines and thus trick geom_text_repel into staying away from them. The complication is that you have to add several filler rows to your data set and then modify the plot to render the filler as invisible. But the end result is pretty clean:
dat2 <- bind_rows(
data.frame(wt = seq(min(dat$wt), max(dat$wt), length = 20), mpg = 19.64, car = 'O'),
data.frame(mpg = seq(min(dat$mpg), max(dat$mpg), length = 20), wt = 3.216, car = 'O'),
dat
)
ggplot(dat2, aes(wt, mpg, label = car))
geom_point(data = filter(dat2, car != 'O'), color = "red")
geom_text_repel(aes(color = car == 'O'), seed = 1, show.legend = F) #Seed for reproducibility
geom_vline(xintercept = 3.216) #Deliberately chosen "bad" numbers
geom_hline(yintercept = 19.64)
scale_color_manual(values = c('black', 'transparent'))
theme_bw()
CodePudding user response:
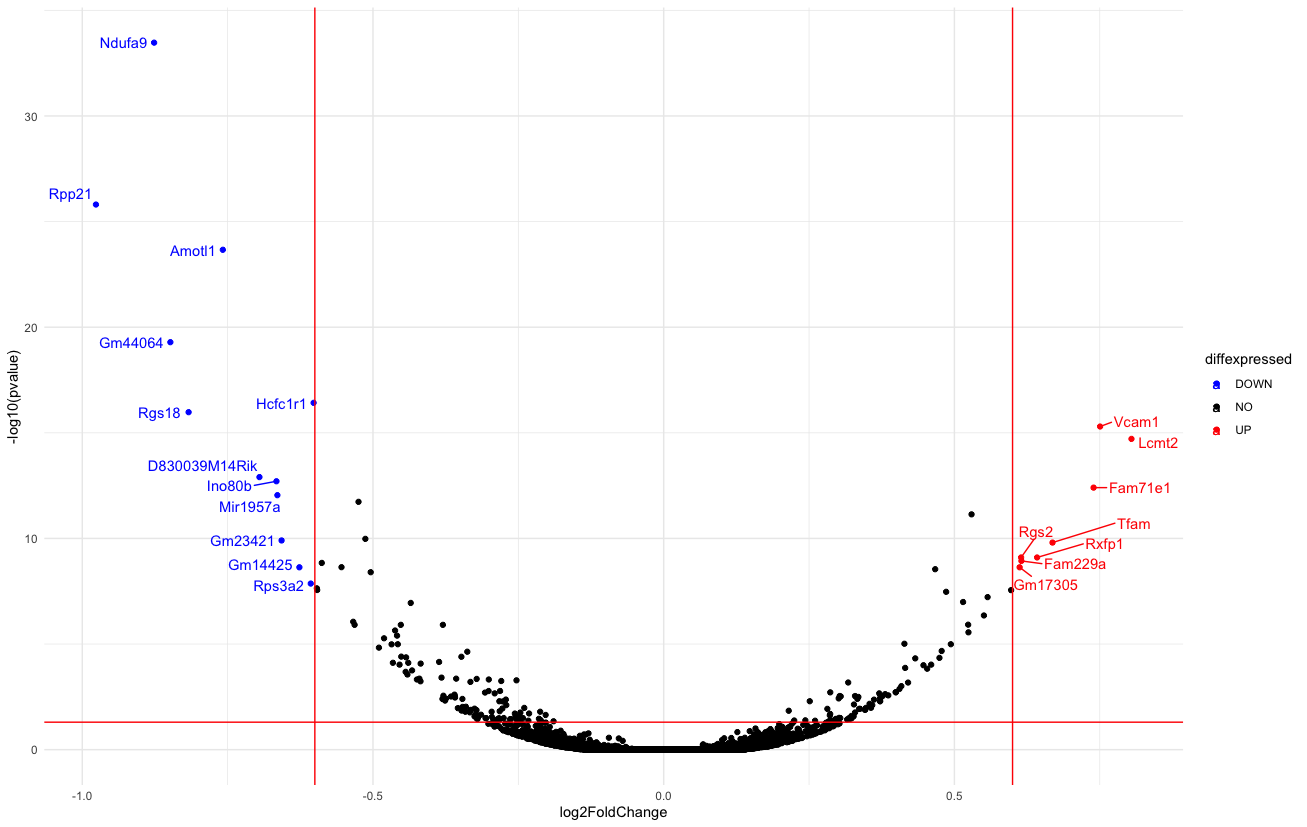
I'm not sure if there's any functions that allows ggrepel to do this automatically. One way to hack around this is to create multiple subsets of data, and add nudge to the label. Here I used the volcano plot as an example.
library(ggplot2)
library(ggrepel)
ggplot(data=de, aes(x=log2FoldChange, y=-log10(pvalue), col=diffexpressed, label=delabel))
geom_point()
theme_minimal()
geom_text_repel(data = subset(de, log2FoldChange < -0.6),
nudge_x = -0.05)
geom_text_repel(data = subset(de, log2FoldChange > 0.6),
nudge_x = 0.08)
scale_color_manual(values=c("blue", "black", "red"))
geom_vline(xintercept=c(-0.6, 0.6), col="red")
geom_hline(yintercept=-log10(0.05), col="red")