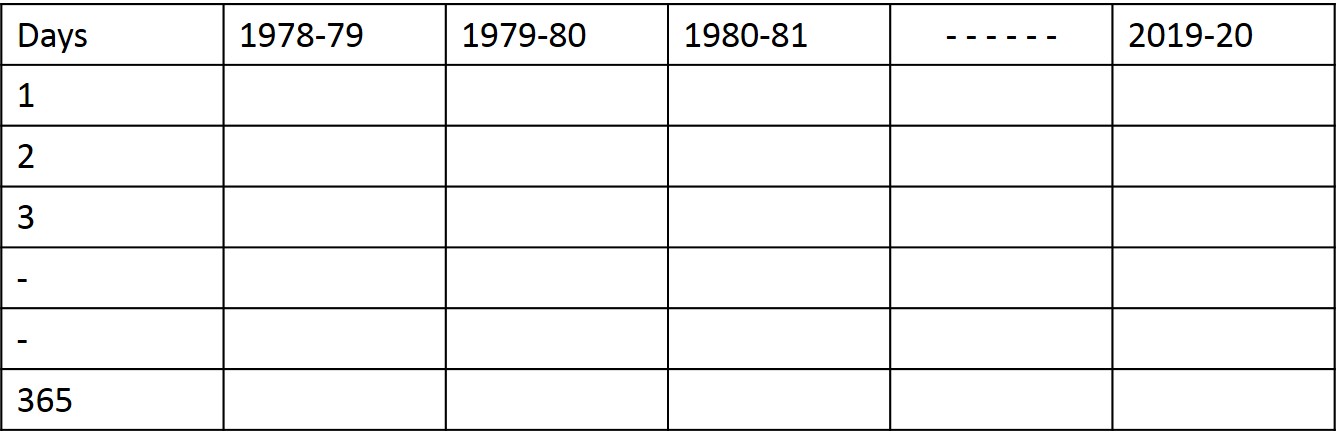
I have daily rainfall data for 43 years. The data is in DD/MM/YYYY format. The start date is 01/01/1978 and the end date is 31/12/2020. I want to reshape/rearrange this data into a table (365 (rows - number of days in a year) x 43 (columns - years of data record)) such that the first day of each data year starts on April 1st and ends on March 31st of the next year (for example, the data year 1978-1979 start from 01/04/1978 and end on 31/03/1979).
Expected table format is:
How to write R code for this? I'm new to R.
CodePudding user response:
This code is admittedly ugly so I am more than willing to take suggestions for improvements.
This solution requires dplyr and lubridate that are both in the tidyverse.
library(lubridate)
library(dplyr)
start_date <- dmy("01/01/1978")
end_date <- dmy("31/12/2020")
# Generate dummy dataset
dates <- as_date(start_date:end_date)
observations <- runif(length(dates), 1, 13)
df <- tibble(dates, observations)
df %>%
# Create a column "dyear" for the data year.
mutate(
dyear = paste(
floor(quarter(dates, type = "year.quarter", fiscal_start = 4)) - 1,
ceiling(quarter(dates, type = "year.quarter", fiscal_start = 4)) - 1,
sep = "-"
)
) %>%
group_by(dyear) %>%
# Hacky method to get the right numbers for day of year.
mutate(yday = if_else(
dyear == "1977-1978",
as.integer(yday(dates) (interval(dmy("01/04/1977"), start_date) %/% days(1))),
row_number()
)) %>%
select(-dates) %>%
pivot_wider(names_from = dyear, values_from = observations, id_expand = T)
I wish yday and year had a fiscal_start option, but it seems that won't be the case.
Either way the code outputs the following into the terminal:
# A tibble: 366 × 45
yday `1977-1978` `1978-1979` `1979-1980` `1980-1981` `1981-1982` `1982-1983`
<int> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 1 NA 9.23 5.86 5.63 4.21 2.14
2 2 NA 9.10 3.20 1.58 6.92 6.33
3 3 NA 5.57 3.05 5.55 6.46 2.91
4 4 NA 5.28 7.62 7.49 13.0 11.7
5 5 NA 7.87 1.58 9.23 10.3 9.67
6 6 NA 8.00 1.13 2.34 3.30 9.02
7 7 NA 12.5 4.98 10.3 2.09 7.13
8 8 NA 10.6 1.86 12.7 10.3 7.40
9 9 NA 7.79 2.76 6.23 7.98 3.15
10 10 NA 3.83 1.21 3.99 2.02 2.94
# … with 356 more rows, and 38 more variables: `1983-1984` <dbl>, `1984-1985` <dbl>,
# `1985-1986` <dbl>, `1986-1987` <dbl>, `1987-1988` <dbl>, `1988-1989` <dbl>,
# `1989-1990` <dbl>, `1990-1991` <dbl>, `1991-1992` <dbl>, `1992-1993` <dbl>,
# `1993-1994` <dbl>, `1994-1995` <dbl>, `1995-1996` <dbl>, `1996-1997` <dbl>,
# `1997-1998` <dbl>, `1998-1999` <dbl>, `1999-2000` <dbl>, `2000-2001` <dbl>,
# `2001-2002` <dbl>, `2002-2003` <dbl>, `2003-2004` <dbl>, `2004-2005` <dbl>,
# `2005-2006` <dbl>, `2006-2007` <dbl>, `2007-2008` <dbl>, `2008-2009` <dbl>, …
Where the NA's are introduced where the data does not exist.
You can pipe this to tail() to see that we get:
# A tibble: 6 × 45
yday `1977-1978` `1978-1979` `1979-1980` `1980-1981` `1981-1982` `1982-1983`
<int> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 361 3.42 6.73 7.90 3.51 9.99 4.43
2 362 3.38 7.77 10.2 9.08 2.63 4.61
3 363 6.65 6.35 2.14 8.16 10.8 8.10
4 364 4.32 8.29 4.68 6.64 10.3 13.0
5 365 3.35 8.06 4.74 9.93 6.19 12.2
6 366 NA NA 12.9 NA NA NA
# … with 38 more variables: `1983-1984` <dbl>, `1984-1985` <dbl>, `1985-1986` <dbl>,
# `1986-1987` <dbl>, `1987-1988` <dbl>, `1988-1989` <dbl>, `1989-1990` <dbl>,
# `1990-1991` <dbl>, `1991-1992` <dbl>, `1992-1993` <dbl>, `1993-1994` <dbl>,
# `1994-1995` <dbl>, `1995-1996` <dbl>, `1996-1997` <dbl>, `1997-1998` <dbl>,
# `1998-1999` <dbl>, `1999-2000` <dbl>, `2000-2001` <dbl>, `2001-2002` <dbl>,
# `2002-2003` <dbl>, `2003-2004` <dbl>, `2004-2005` <dbl>, `2005-2006` <dbl>,
# `2006-2007` <dbl>, `2007-2008` <dbl>, `2008-2009` <dbl>, `2009-2010` <dbl>, …
Which shows which data years contain the extra day.
CodePudding user response:
You may simply split rainfall data at the cumsum of '04-01'. This gives a list S with 365 or 366 measures. Just adapt all `length<-` to 366, where NA is added in case. Finally just cbind and add day column. The column names for the years can be generated using strftime.
At first we need to expand the missing start and end, i.e. April 1 before your measures start and March 31 after, and merge it to the data.
aux <- data.frame(
date=seq.Date(as.Date("1977-04-01"), as.Date("1982-03-31"), 'day')
)
dat <- merge(dat, aux, all=TRUE)
Solution 1, more rows for leap years
S <- split(dat$rainfall, cumsum(strftime(dat$date, '%m-%d') == '04-01'))
yrs <- unique(strftime(dat$date, '%Y'))
yrs <- apply(cbind(yrs[-length(yrs)], yrs[-1]), 1, \(x)
paste(x[[1]], substr(x[[2]], 3, 4), sep='_'))
res1 <- cbind(day=1:366, setNames(do.call(cbind.data.frame, lapply(S, `length<-`, 366)),
yrs))
Gives:
tail(res1)
# day 1977_78 1978_79 1979_80 1980_81 1981_82
# 361 361 81.199690 110.382115 75.45856 134.532854 NA
# 362 362 1.713985 5.091447 13.14269 7.704723 NA
# 363 363 1.724329 37.100737 96.13059 42.934299 NA
# 364 364 113.334633 20.346748 97.99042 73.300045 NA
# 365 365 102.177311 30.465944 341.11990 133.788648 NA
# 366 366 NA NA 235.38249 NA NA
Solution 2, all rows 366
If we want the days to be aligned through the years, we could append an NA after day 334 in non-leap years.
r <- by(dat, cumsum(strftime(dat$date, '%m-%d') == '04-01'), \(x) {
rf <- x$rainfall
yrs <- unique(strftime(x$date, '%Y'))
yrs <- paste(yrs[1], substr(yrs[2], 3, 4), sep='_')
if (length(rf) != 366) {
rf <- append(rf, NA, 334)
}
`attr<-`(rf, 'yrs', yrs)
})
res2 <- cbind(day=1:366, setNames(do.call(cbind.data.frame, r), sapply(r, attr, 'yrs')))
Gives
res2[331:337, ]
# day 1977_78 1978_79 1979_80 1980_81 1981_82
# 331 331 47.65507 99.59559 38.80979 126.29785 NA
# 332 332 87.92888 125.35296 46.02958 82.03465 NA
# 333 333 28.98329 29.83954 31.30135 87.30667 NA
# 334 334 279.30901 39.37710 12.03988 283.37096 NA
# 335 335 NA NA 206.63815 NA NA
# 336 336 48.48830 108.82812 45.95315 36.61144 NA
# 337 337 16.72346 25.30704 36.15601 210.51755 NA
Data:
dat <- data.frame(
date=seq.Date(as.Date('1978-01-01'), as.Date('1981-12-31'), 'day')
)
set.seed(42)
dat$rainfall <- rnorm(nrow(dat), 20, 100) |> abs()