Suppose I have the following data.frame object:
df = data.frame(id=(1:25),
col1=c('a','a','a',
'b','b','b',
'c','c','c',
'd','d',
'b','b','b',
'e',
'c','c','c',
'e','e',
'a','a','a',
'e','e'))
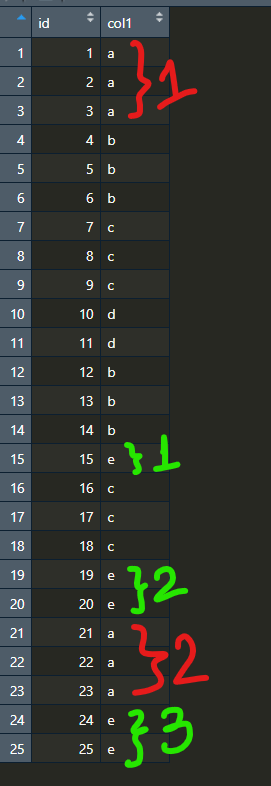
From the snapshot above, you can see that there are two groups of rows that have col1=="a": rows 1 through 3 and rows 21 through 23. Similarly, there are three groups of rows that have col1=="e": row 15, rows 19 through 20 and rows 24 through 25 (and so on and so on with "b", "c" and "d").
Here's my main question
Is it possible to label the rows according to what "chunk" we're currently on? More explicitly: since rows 1 through 3 are the first time where we have col1=="a", they should be labelled as 1. Then, rows 21 through 23 should be labelled as 2, because that is the second time that we have a set of rows that have col1=="a". Using the same logic, but for col1=="e", we'd label row 15 as 1, rows 19 and 20 as 2 and rows 24 and 25 as 3 (again, so on and so on with "b", "c" and "d").
Desired output
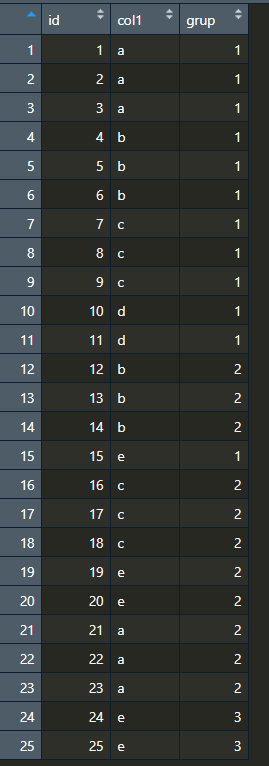
Here is what the resulting data.frame would look like:
df = data.frame(id=(1:25),
col1=c('a','a','a',
'b','b','b',
'c','c','c',
'd','d',
'b','b','b',
'e',
'c','c','c',
'e','e',
'a','a','a',
'e','e'),
grup=c(1,1,1,
1,1,1,
1,1,1,
1,1,
2,2,2,
1,
2,2,2,
2,2,
2,2,2,
3,3))
My attempt
I tried implementing a solution using a for loop, but that was quite slow (the original data I'm working on has about 500,000 rows), and it just looked a bit sloppy:
my_classifier = function(input_df, ref_column){
# Keeps a tally of how many times each unique group was "found" before.
group_counter = list()
# Dealing with the corner case of the first row
group_counter[[df$col1[1]]] = 1
output_groups = rep(-1, nrow(input_df))
output_groups[1] = 1
# The for loop starts at the second row because I've already "dealt" with the
# first row in the corner cases above
for(i in 2:nrow(input_df)){
prev_group = input_df[[ref_column]][i-1]
this_group = input_df[[ref_column]][i]
if(is.null(group_counter[[this_group]])){
this_counter = 0
}
else{
this_counter = group_counter[[this_group]]
}
if(prev_group != this_group){
this_counter = this_counter 1
}
output_groups[i] = this_counter
group_counter[[this_group]] = this_counter
}
return(output_groups)
}
df$grup = my_classifier(df,'col1')
Is there a quicker/more efficient way to solve this problem? Maybe something that relies on vectorized functions or something?
Important notes
Consider that we cannot rely on the number of repetitions of each "block". Sometimes, col1 will have just one single row of a particular group, while other times the "block" will have several rows where col1 share the same value. Also consider that we cannot assume any logic in the "order" or the number of times each group shows up.
So, for example, there might be a a stretch of 10 rows where col1=="z", then a stretch of 15 rows where col1=="x", then another single row where col1=="x" and then finally a stretch of 100 rows where col1=="w".
CodePudding user response:
You can use data.table::rleid() twice, like this:
library(data.table)
setDT(df)[,grp:=rleid(col1)][, grp:=rleid(grp), by=col1][order(id)]
Output:
id col1 grp
<int> <char> <int>
1: 1 a 1
2: 2 a 1
3: 3 a 1
4: 4 b 1
5: 5 b 1
6: 6 b 1
7: 7 c 1
8: 8 c 1
9: 9 c 1
10: 10 d 1
11: 11 d 1
12: 12 b 2
13: 13 b 2
14: 14 b 2
15: 15 e 1
16: 16 c 2
17: 17 c 2
18: 18 c 2
19: 19 e 2
20: 20 e 2
21: 21 a 2
22: 22 a 2
23: 23 a 2
24: 24 e 3
25: 25 e 3
id col1 grp
CodePudding user response:
Here is a possible base R solution:
change <- with(rle(df$col1), rep(seq_along(values), lengths))
cbind(df, grp = with(df, ave(
change,
col1,
FUN = function(x)
inverse.rle(within.list(rle(x), values <- seq_along(values)))
)))
Or another option using a combination of rle and dplyr using the function from here:
rle_new <- function(x) {
x <- rle(x)$lengths
rep(seq_along(x), times=x)
}
library(dplyr)
df %>%
mutate(grp = rle_new(col1)) %>%
group_by(col1) %>%
mutate(grp = rle_new(grp))
Output
id col1 grp
1 1 a 1
2 2 a 1
3 3 a 1
4 4 b 1
5 5 b 1
6 6 b 1
7 7 c 1
8 8 c 1
9 9 c 1
10 10 d 1
11 11 d 1
12 12 b 2
13 13 b 2
14 14 b 2
15 15 e 1
16 16 c 2
17 17 c 2
18 18 c 2
19 19 e 2
20 20 e 2
21 21 a 2
22 22 a 2
23 23 a 2
24 24 e 3
25 25 e 3