DATA = data.frame("GROUP" = sort(rep(1:4, 200)),
"TYPE" = rep(1:2, 400),
"TIME" = rep(100:101, 400),
"SCORE" = sample(1:100,r=T,800))
Cheers all,
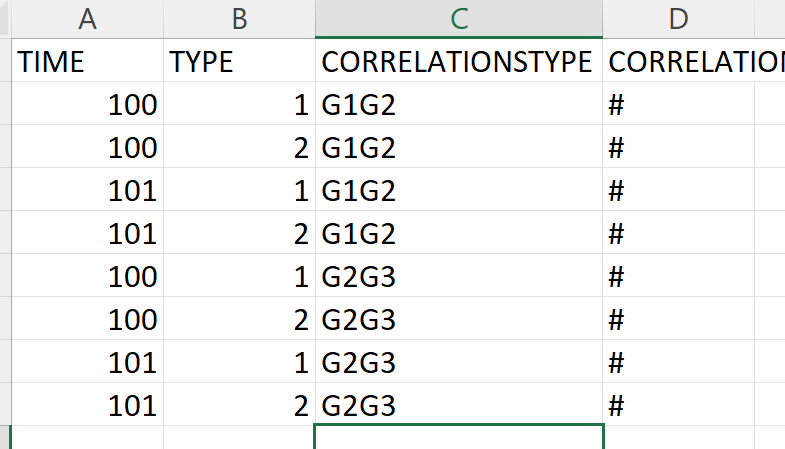
I have 'DATA' and wish to estimation the CORRELATION VALUES of SCORE at each TIME and SCORE and TYPE combination BETWEEN AND WITHIN GROUP in this way:
CodePudding user response:
I am assuming you want to compute the correlation between groups 1-2, 1-3, 1-4 and so on for each combination of TIME and TYPE. Here's an approach:
# create the dataset
set.seed(123)
df <- data.frame("group" = sort(rep(1:4, 200)),
"type" = rep(1:2, 400),
"time" = rep(100:101, 400),
"score" = sample(1:100,r=T,800))
library(tidyr)
library(purrr)
library(data.table)
# another dataset to filter combinations
# (G1G2 is same G2G1, so remove G2G1)
df2 <- combn(4, 2) %>% t %>%
as_tibble() %>%
rename(group1 = V1, group2 = V2) %>%
mutate(value = TRUE)
df %>%
# add identifiers per group
group_by(time, type, group) %>%
mutate(id = row_number()) %>%
ungroup() %>%
# nest data to get separate tibble for each
# combination of time and type
nest(data = -c(time, type)) %>%
# convert each data.frame to data.table
mutate(dt = map(data, function(dt){
setDT(dt)
setkey(dt, id)
dt
})) %>%
# correlation between groups in R
# refer answer below for more details
# https://stackoverflow.com/a/26357667/15221658
# cartesian join of dts
mutate(dtj = map(dt, ~.[., allow.cartesian = TRUE])) %>%
# compute between group correlation
mutate(cors = map(dtj, ~.[, list(cors = cor(score, i.score)), by = list(group, i.group)])) %>%
# unnest correlation object
unnest(cors) %>%
# formatting for display
select(type, time, group1 = group, group2 = i.group, correlation = cors) %>%
filter(group1 != group2) %>%
arrange(time, group1, group2) %>%
# now use df2 since currently we have G1G2, and G2G1
# which are both equal so remove G2G1
left_join(df2, by = c("group1", "group2")) %>%
filter(value) %>%
select(-value)
# A tibble: 12 x 5
type time group1 group2 correlation
<int> <int> <int> <int> <dbl>
1 1 100 1 2 0.121
2 1 100 1 3 0.0543
3 1 100 1 4 -0.0694
4 1 100 2 3 -0.104
5 1 100 2 4 -0.0479
6 1 100 3 4 -0.0365
7 2 101 1 2 -0.181
8 2 101 1 3 -0.0673
9 2 101 1 4 0.00765
10 2 101 2 3 0.0904
11 2 101 2 4 -0.0126
12 2 101 3 4 -0.154
CodePudding user response:
Here is an alternative approach:
library(data.table)
setDT(DATA, key = c("GROUP", "TYPE", "TIME"))[
, CJ(TY = TYPE, TI = TIME, GA = GROUP, GB = GROUP, unique = TRUE)][GA < GB][
, CORRELATIONSTYPE := paste0("G", GA, "G", GB)][
, CORRELATION := cor(DATA[.(GA, TY, TI), SCORE], DATA[.(GB, TY, TI), SCORE]), by = .I][]
Key: <TY, TI, GA, GB> TY TI GA GB CORRELATIONSTYPE CORRELATION <int> <int> <int> <int> <char> <num> 1: 1 100 1 2 G1G2 0.12240303 2: 1 100 1 3 G1G3 -0.03969913 3: 1 100 1 4 G1G4 -0.09666560 4: 1 100 2 3 G2G3 -0.02250806 5: 1 100 2 4 G2G4 -0.07106922 6: 1 100 3 4 G3G4 -0.13140392 7: 1 101 1 2 G1G2 NA 8: 1 101 1 3 G1G3 NA 9: 1 101 1 4 G1G4 NA 10: 1 101 2 3 G2G3 NA 11: 1 101 2 4 G2G4 NA 12: 1 101 3 4 G3G4 NA 13: 2 100 1 2 G1G2 NA 14: 2 100 1 3 G1G3 NA 15: 2 100 1 4 G1G4 NA 16: 2 100 2 3 G2G3 NA 17: 2 100 2 4 G2G4 NA 18: 2 100 3 4 G3G4 NA 19: 2 101 1 2 G1G2 -0.03600460 20: 2 101 1 3 G1G3 0.03328082 21: 2 101 1 4 G1G4 -0.09641611 22: 2 101 2 3 G2G3 -0.06334729 23: 2 101 2 4 G2G4 0.06455382 24: 2 101 3 4 G3G4 -0.07121206 TY TI GA GB CORRELATIONSTYPE CORRELATION
