I have a dataframe in R where each row is a song, and each column is an artist that collaborates on the song. I want to visualise which artists work together. I'm figuring I could do some kind of network analysis, but I don't know how to convert my dataframe to a network, or something with nodes and edges, or a vertex.
song <- c('You','Strings','Club','Never','Yolo','Do it','Again','The world', 'Forever')
artist1 <- c('Art', 'Bee', 'Con', 'Con', 'Dev', 'Art', 'Evi', 'Evi', 'Guy')
artist2 <- c('Bee', 'Evi', NA, 'Bee', 'Fay', 'Con', 'Bee', 'Dev', NA)
artist3 <- c('Con', 'Fay', NA, NA, NA, 'Bee', 'Art', 'Con', NA)
df <- data.frame(song, artist1, artist2, artist3)
What I guess I need to count (and then visualise) is how often each artist occurs in the same row as each other artist. The network would then show the artists as nodes and the collaborations (weighted) as edges.
Any clues as to how to go about?
CodePudding user response:
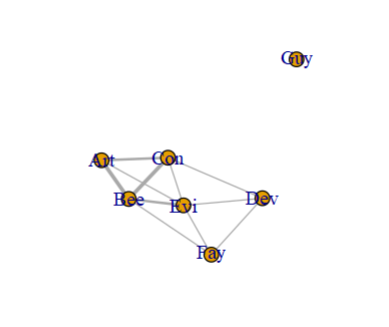
Create an adjacency matrix with crossprod, then use graph_from_adjacency_matrix to create a graph object, and then plot it:
ta <- crossprod(table(cbind(df[1], stack(df[-1]))[-3]))
graph_from_adjacency_matrix(ta, diag = F, weighted = T, mode = "undirected") %>%
plot(edge.width = E(.)$weight)
CodePudding user response:
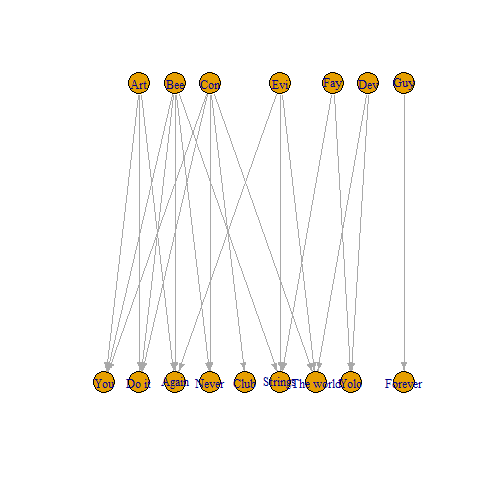
You can try the code below
df %>%
pivot_longer(!song, values_drop_na = TRUE) %>%
select(!name) %>%
rev() %>%
graph_from_data_frame() %>%
set_vertex_attr(name = "type", value = names(V(.)) %in% df$song) %>%
plot(layout = layout_as_bipartite)