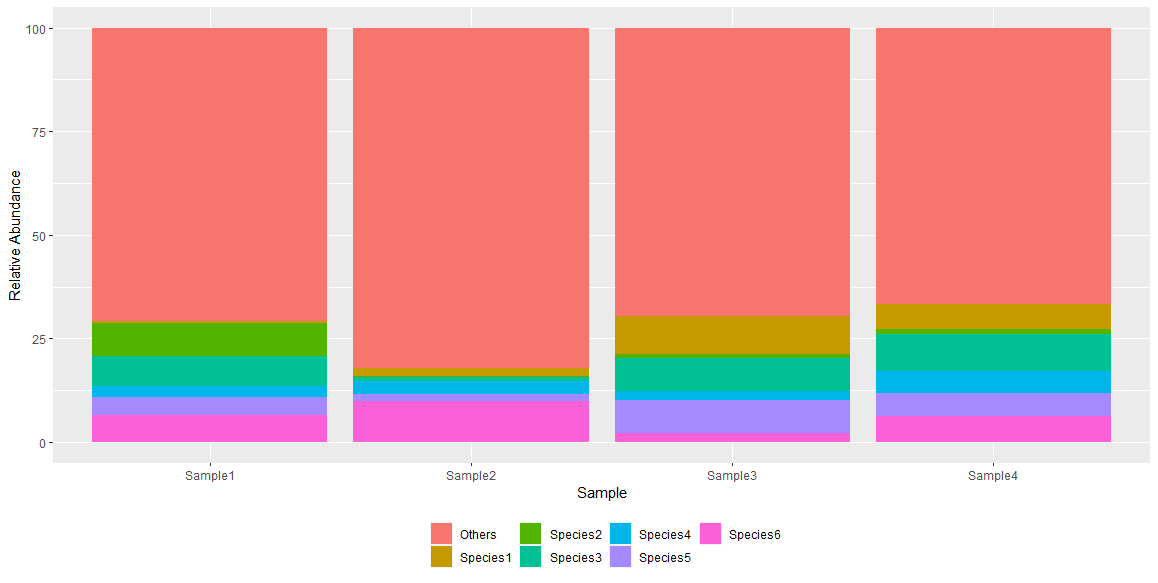
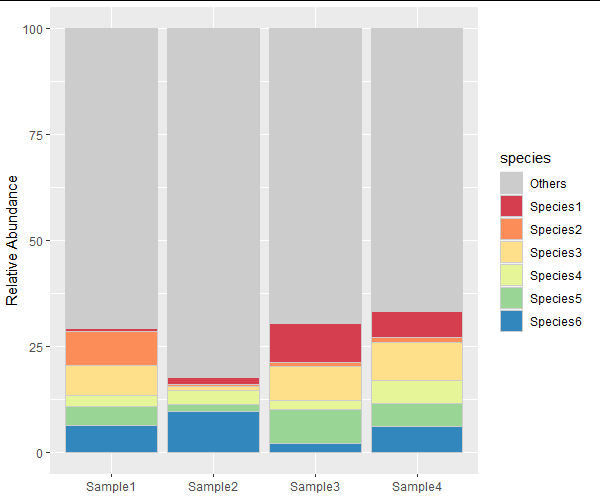
I have a multivariate dataset of species relative abundance (%) in different samples. In this data frame I only have the most abundant species, so the total is not 100%.
My dataset looks like this but with much more species and samples:
species = c("Species1","Species2","Species3","Species4","Species5","Species6")
Sample1 = c(0.6,7.9,7.1,2.7,4.5,6.4)
Sample2 = c(1.8,0.3,0.9,3.3,1.7,9.8)
Sample3 = c(9.2,1,8,2.1,8,2.2)
Sample4 = c(6.1,1.3,9,5.3,5.5,6.2)
df = data.frame(species, Sample1, Sample2, Sample3, Sample4)
df
species Sample1 Sample2 Sample3 Sample4
1 Species1 0.6 1.8 9.2 6.1
2 Species2 7.9 0.3 1.0 1.3
3 Species3 7.1 0.9 8.0 9.0
4 Species4 2.7 3.3 2.1 5.3
5 Species5 4.5 1.7 8.0 5.5
6 Species6 6.4 9.8 2.2 6.2
But I would like to make a stacked bar plot where I also have the variable "Others" representing the percentage cover of all the rarest species together, calculated as 100 - sum of column
The result I would like is this:
species Sample1 Sample2 Sample3 Sample4
1 Species1 0.6 1.8 9.2 6.1
2 Species2 7.9 0.3 1.0 1.3
3 Species3 7.1 0.9 8.0 9.0
4 Species4 2.7 3.3 2.1 5.3
5 Species5 4.5 1.7 8.0 5.5
6 Species6 6.4 9.8 2.2 6.2
7 Others 70.8 82.2 69.5 66.6
How can I do? I've been searching for hours but I couldn't find a solution.
CodePudding user response:
To get the data you need, use summarize(across())
bind_rows(
df,
df %>% summarize(across(starts_with("Sample"),~100-sum(.x))) %>%
mutate(species="Others")
)
Output:
species Sample1 Sample2 Sample3 Sample4
1 Species1 0.6 1.8 9.2 6.1
2 Species2 7.9 0.3 1.0 1.3
3 Species3 7.1 0.9 8.0 9.0
4 Species4 2.7 3.3 2.1 5.3
5 Species5 4.5 1.7 8.0 5.5
6 Species6 6.4 9.8 2.2 6.2
7 Others 70.8 82.2 69.5 66.6
Furthermore, if you want to plot this in a simple stacked bar chart, you can continue the pipepline with this:
... %>% pivot_longer(cols = -species, names_to="Sample",values_to = "Abundance") %>%
ggplot(aes(Sample,Abundance,fill=species))
geom_col()
labs(fill="", y="Relative Abundance")
theme(legend.position = "bottom")
CodePudding user response:
The main difference btw this and the other answer is the use of data.table:
library(data.table)
library(ggplot2)
library(RColorBrewer)
#
setDT(df)
result <- rbind(df, df[, c(species='Others', lapply(.SD, \(x) 100-sum(x))), .SDcols=-1])
result
## species Sample1 Sample2 Sample3 Sample4
## 1: Species1 0.6 1.8 9.2 6.1
## 2: Species2 7.9 0.3 1.0 1.3
## 3: Species3 7.1 0.9 8.0 9.0
## 4: Species4 2.7 3.3 2.1 5.3
## 5: Species5 4.5 1.7 8.0 5.5
## 6: Species6 6.4 9.8 2.2 6.2
## 7: Others 70.8 82.2 69.5 66.6
.SDcols = -1 means use all columns but the first one.
# melt for use in ggplot
# reorder factors to put "Others" at the top.
#
gg.dt <- melt(result, id='species')[
, species:=factor(species, levels=c('Others', setdiff(unique(species), 'Others')))]
##
# use Brewer palette, with grey80 for "Others"
#
ggplot(gg.dt, aes(x=variable, y=value, fill=species))
geom_bar(stat='identity', color='grey80')
scale_fill_manual(values = c('grey80', brewer.pal(6, 'Spectral')))
labs(x=NULL, y='Relative Abundance')
CodePudding user response:
Another possible solution, based on dplyr and colSums:
library(dplyr)
df %>%
bind_rows(data.frame(species = "Others", t(100 - colSums(.[-1]))))
#> species Sample1 Sample2 Sample3 Sample4
#> 1 Species1 0.6 1.8 9.2 6.1
#> 2 Species2 7.9 0.3 1.0 1.3
#> 3 Species3 7.1 0.9 8.0 9.0
#> 4 Species4 2.7 3.3 2.1 5.3
#> 5 Species5 4.5 1.7 8.0 5.5
#> 6 Species6 6.4 9.8 2.2 6.2
#> 7 Others 70.8 82.2 69.5 66.6