(This is my first StackOverflow question ever!)
I have a pandas dataframe that contains solar irradiance values in 15-minute intervals over the course of a single day. This dataframe's index is a "DatetimeIndex" (dtype='datetime64[ns, America/New_York]', and is localized to its respective timezone. So, the index values start as "1997-01-20 00:15:00-05:00", "1997-01-20 00:30:00-05:00" ... "1997-01-21 00:00:00-5:00" (notice the last entry is 12AM of the next day).
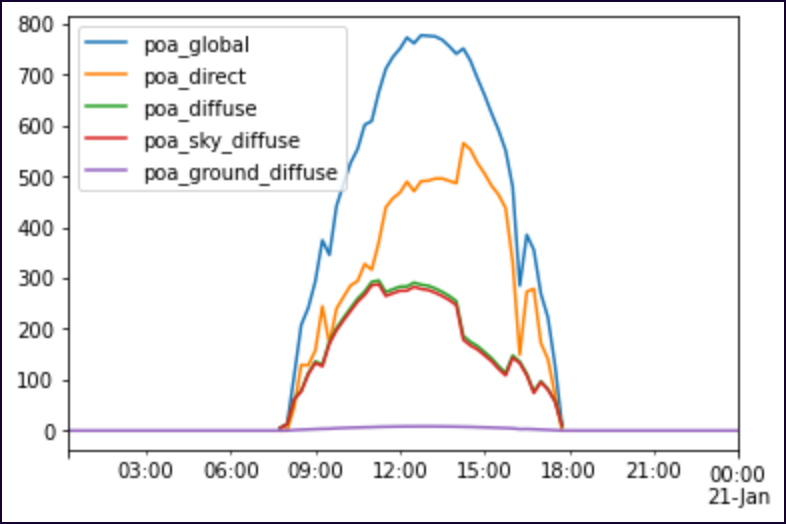
When plotting this dataframe (named poa_irradiance_swd_corrected) by itself, everything looks great. Here's an example of my code and its output:
Code:
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
from matplotlib.dates import DateFormatter
import pvlib as pv
import datetime as dt
import math
import pytz
import time
plt.figure()
poa_irradiance_swd_corrected.plot().get_figure().set_facecolor('white')
The highest "poa_global" irradiance measurements in this dataframe are between 12:00 PM (noon) and 1:30-ish. This is accurate.
I also have two other dataframes (named poa_irradiance_swd_flat and modeled_poa_irradiance_swd) with similar irradiance measurements. To compare the irradiance measurements visually, I put all 3 dataframes into a single figure using matplotlib's subplot function. However, the X-axis of the subplots are very inaccurate. The code and output are shown below:
Code:
fig, axs = plt.subplots(2,2, sharex=True,sharey=True, facecolor='white',figsize=(12,8))
fig.suptitle('Sunny Winter Day',size='xx-large',weight='bold')
axs[0,0].plot(poa_irradiance_swd_flat)
axs[0,0].set_title('POA Irradiance (Flat)')
axs[0,1].plot(modeled_poa_irradiance_swd)
axs[0,1].set_title('Modeled Data (Tilted)')
axs[1,0].plot(poa_irradiance_swd_corrected)
axs[1,0].set_title('POA Irradiance (Tilted)')
axs[1,1].plot(modeled_poa_irradiance_swd)
axs[1,1].set_title('Modeled Data (Tilted)')
for axs in axs.flat:
axs.set(ylabel='Irradiance $W/m^2$')
axs.xaxis.set_major_formatter(DateFormatter('%H:%M'))
plt.tight_layout(pad=0, w_pad=0, h_pad=3)
fig.autofmt_xdate()
Output: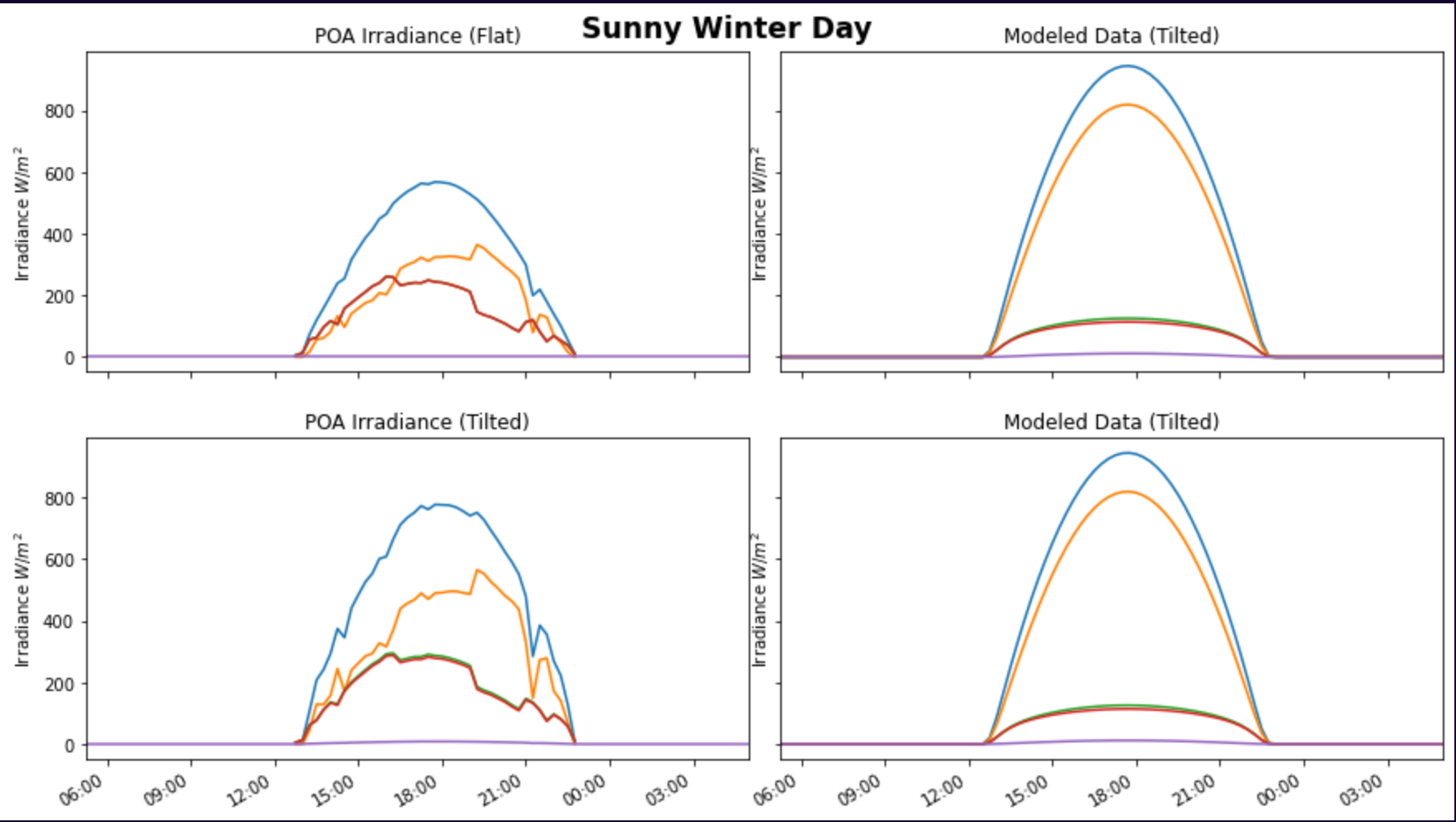 Note that the two graphs on the right are the exact same. I simply used the graphs to the right to compare to the graphs to the left
Note that the two graphs on the right are the exact same. I simply used the graphs to the right to compare to the graphs to the left
Notice how the x-axis is completely off by 6-ish hours.
I've cross-checked the dataframes themselves, and again, the highest irradiance times coincide with the times between 12:00 PM (noon) and around 1:30-ish... not around 18:00 (which is 6:00 PM and does not make sense). I even used the same exact dataframe in the axs[1,0].plot(poa_irradiance_swd_corrected) line of code.
How can I get the X-axis to display the accurate times?
EDIT (in response to scespinoza):
So I tried passing the respective .index and .values values from the dataframe, and it successfully plotted... but still with the same problem.
Then, I tried passing the ax commands into the plot function itself like this: poa_irradiance_swd_flat.plot(ax=axs[0, 0]) for each subplot. After importing matplotlib as mpl, I also added mpl.rcParams['timezone'] = 'UTC' to the top of my code.
Now, I'm getting a ValueError:
ValueError Traceback (most recent call last)
File ~/opt/miniconda3/envs/pvlib/lib/python3.10/site-packages/IPython/core/formatters.py:339, in BaseFormatter.__call__(self, obj)
337 pass
338 else:
--> 339 return printer(obj)
340 # Finally look for special method names
341 method = get_real_method(obj, self.print_method)
File ~/opt/miniconda3/envs/pvlib/lib/python3.10/site-packages/IPython/core/pylabtools.py:151, in print_figure(fig, fmt, bbox_inches, base64, **kwargs)
148 from matplotlib.backend_bases import FigureCanvasBase
149 FigureCanvasBase(fig)
--> 151 fig.canvas.print_figure(bytes_io, **kw)
152 data = bytes_io.getvalue()
153 if fmt == 'svg':
File ~/opt/miniconda3/envs/pvlib/lib/python3.10/site-packages/matplotlib/backend_bases.py:2295, in FigureCanvasBase.print_figure(self, filename, dpi, facecolor, edgecolor, orientation, format, bbox_inches, pad_inches, bbox_extra_artists, backend, **kwargs)
2289 renderer = _get_renderer(
2290 self.figure,
2291 functools.partial(
2292 print_method, orientation=orientation)
2293 )
2294 with getattr(renderer, "_draw_disabled", nullcontext)():
-> 2295 self.figure.draw(renderer)
2297 if bbox_inches:
2298 if bbox_inches == "tight":
File ~/opt/miniconda3/envs/pvlib/lib/python3.10/site-packages/matplotlib/artist.py:73, in _finalize_rasterization.<locals>.draw_wrapper(artist, renderer, *args, **kwargs)
71 @wraps(draw)
72 def draw_wrapper(artist, renderer, *args, **kwargs):
---> 73 result = draw(artist, renderer, *args, **kwargs)
74 if renderer._rasterizing:
75 renderer.stop_rasterizing()
File ~/opt/miniconda3/envs/pvlib/lib/python3.10/site-packages/matplotlib/artist.py:50, in allow_rasterization.<locals>.draw_wrapper(artist, renderer)
47 if artist.get_agg_filter() is not None:
48 renderer.start_filter()
---> 50 return draw(artist, renderer)
51 finally:
52 if artist.get_agg_filter() is not None:
File ~/opt/miniconda3/envs/pvlib/lib/python3.10/site-packages/matplotlib/figure.py:2810, in Figure.draw(self, renderer)
2807 # ValueError can occur when resizing a window.
2809 self.patch.draw(renderer)
-> 2810 mimage._draw_list_compositing_images(
2811 renderer, self, artists, self.suppressComposite)
2813 for sfig in self.subfigs:
2814 sfig.draw(renderer)
File ~/opt/miniconda3/envs/pvlib/lib/python3.10/site-packages/matplotlib/image.py:132, in _draw_list_compositing_images(renderer, parent, artists, suppress_composite)
130 if not_composite or not has_images:
131 for a in artists:
--> 132 a.draw(renderer)
133 else:
134 # Composite any adjacent images together
135 image_group = []
File ~/opt/miniconda3/envs/pvlib/lib/python3.10/site-packages/matplotlib/artist.py:50, in allow_rasterization.<locals>.draw_wrapper(artist, renderer)
47 if artist.get_agg_filter() is not None:
48 renderer.start_filter()
---> 50 return draw(artist, renderer)
51 finally:
52 if artist.get_agg_filter() is not None:
File ~/opt/miniconda3/envs/pvlib/lib/python3.10/site-packages/matplotlib/axes/_base.py:3046, in _AxesBase.draw(self, renderer)
3043 for spine in self.spines.values():
3044 artists.remove(spine)
-> 3046 self._update_title_position(renderer)
3048 if not self.axison:
3049 for _axis in self._get_axis_list():
File ~/opt/miniconda3/envs/pvlib/lib/python3.10/site-packages/matplotlib/axes/_base.py:2986, in _AxesBase._update_title_position(self, renderer)
2983 for ax in axs:
2984 if (ax.xaxis.get_ticks_position() in ['top', 'unknown']
2985 or ax.xaxis.get_label_position() == 'top'):
-> 2986 bb = ax.xaxis.get_tightbbox(renderer)
2987 else:
2988 bb = ax.get_window_extent(renderer)
File ~/opt/miniconda3/envs/pvlib/lib/python3.10/site-packages/matplotlib/axis.py:1103, in Axis.get_tightbbox(self, renderer, for_layout_only)
1100 if not self.get_visible():
1101 return
-> 1103 ticks_to_draw = self._update_ticks()
1105 self._update_label_position(renderer)
1107 # go back to just this axis's tick labels
File ~/opt/miniconda3/envs/pvlib/lib/python3.10/site-packages/matplotlib/axis.py:1046, in Axis._update_ticks(self)
1041 """
1042 Update ticks (position and labels) using the current data interval of
1043 the axes. Return the list of ticks that will be drawn.
1044 """
1045 major_locs = self.get_majorticklocs()
-> 1046 major_labels = self.major.formatter.format_ticks(major_locs)
1047 major_ticks = self.get_major_ticks(len(major_locs))
1048 self.major.formatter.set_locs(major_locs)
File ~/opt/miniconda3/envs/pvlib/lib/python3.10/site-packages/matplotlib/ticker.py:224, in Formatter.format_ticks(self, values)
222 """Return the tick labels for all the ticks at once."""
223 self.set_locs(values)
--> 224 return [self(value, i) for i, value in enumerate(values)]
File ~/opt/miniconda3/envs/pvlib/lib/python3.10/site-packages/matplotlib/ticker.py:224, in <listcomp>(.0)
222 """Return the tick labels for all the ticks at once."""
223 self.set_locs(values)
--> 224 return [self(value, i) for i, value in enumerate(values)]
File ~/opt/miniconda3/envs/pvlib/lib/python3.10/site-packages/matplotlib/dates.py:636, in DateFormatter.__call__(self, x, pos)
635 def __call__(self, x, pos=0):
--> 636 result = num2date(x, self.tz).strftime(self.fmt)
637 return _wrap_in_tex(result) if self._usetex else result
File ~/opt/miniconda3/envs/pvlib/lib/python3.10/site-packages/matplotlib/dates.py:528, in num2date(x, tz)
526 if tz is None:
527 tz = _get_rc_timezone()
--> 528 return _from_ordinalf_np_vectorized(x, tz).tolist()
File ~/opt/miniconda3/envs/pvlib/lib/python3.10/site-packages/numpy/lib/function_base.py:2163, in vectorize.__call__(self, *args, **kwargs)
2160 vargs = [args[_i] for _i in inds]
2161 vargs.extend([kwargs[_n] for _n in names])
-> 2163 return self._vectorize_call(func=func, args=vargs)
File ~/opt/miniconda3/envs/pvlib/lib/python3.10/site-packages/numpy/lib/function_base.py:2246, in vectorize._vectorize_call(self, func, args)
2243 # Convert args to object arrays first
2244 inputs = [asanyarray(a, dtype=object) for a in args]
-> 2246 outputs = ufunc(*inputs)
2248 if ufunc.nout == 1:
2249 res = asanyarray(outputs, dtype=otypes[0])
File ~/opt/miniconda3/envs/pvlib/lib/python3.10/site-packages/matplotlib/dates.py:350, in _from_ordinalf(x, tz)
347 dt = (np.datetime64(get_epoch())
348 np.timedelta64(int(np.round(x * MUSECONDS_PER_DAY)), 'us'))
349 if dt < np.datetime64('0001-01-01') or dt >= np.datetime64('10000-01-01'):
--> 350 raise ValueError(f'Date ordinal {x} converts to {dt} (using '
351 f'epoch {get_epoch()}), but Matplotlib dates must be '
352 'between year 0001 and 9999.')
353 # convert from datetime64 to datetime:
354 dt = dt.tolist()
ValueError: Date ordinal 14228655 converts to 40926-09-26T00:00:00.000000 (using epoch 1970-01-01T00:00:00), but Matplotlib dates must be between year 0001 and 9999.
CodePudding user response:
Can't really reproduce your problem, but maybe you can try passing the index and values of the series by separate to the plot function.
...
axs[0,0].plot(poa_irradiance_swd_flat.index, poa_irradiance_swd_flat.values)
...
Also, note that you can pass an ax attribute to the .plot() function of a Series, so you can refactor your code to something like this:
fig, axs = plt.subplots(2,2, sharex=True,sharey=True, facecolor='white',figsize=(12,8))
fig.suptitle('Sunny Winter Day',size='xx-large',weight='bold')
poa_irradiance_swd_flat.plot(ax=axs[0, 0])
axs[0,0].set_title('POA Irradiance (Flat)')
modeled_poa_irradiance_swd.plot(ax=axs[0, 1])
axs[0,1].set_title('Modeled Data (Tilted)')
poa_irradiance_swd_corrected.plot(ax=axs[1, 0])
axs[1,0].set_title('POA Irradiance (Tilted)')
modeled_poa_irradiance_swd.plot(ax=axs[1, 1])
axs[1,1].set_title('Modeled Data (Tilted)')
for axs in axs.flat:
axs.set(ylabel='Irradiance $W/m^2$')
# axs.xaxis.set_major_formatter(DateFormatter('%H:%M'))
plt.tight_layout(pad=0, w_pad=0, h_pad=3)
fig.autofmt_xdate()
Update
I think the error may come from DateFormatter converting the timezone of your date values. Try adding this on the top of your program.
import matplotlib as mpl
mpl.rcParams['timezone'] = 'UTC'