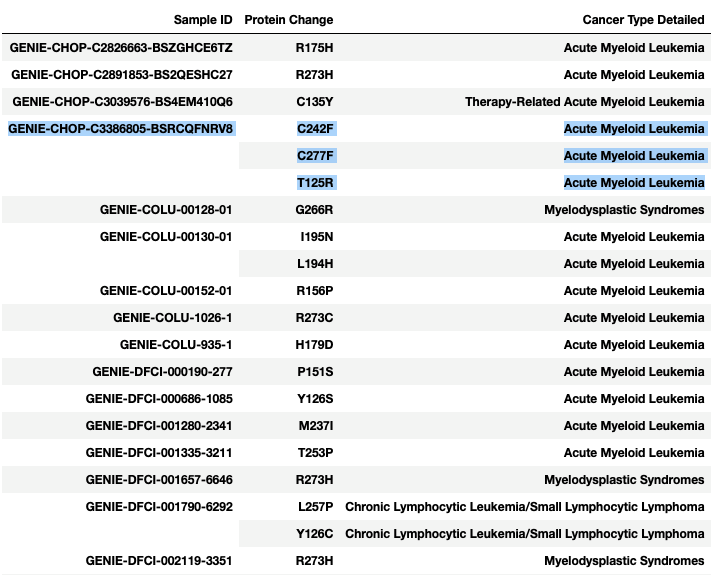
I'm trying to find a nice way to visualize the data from publicly available cBioPortal mutation data. I want to plot the co-occurrence of Protein Change (so basically, for each sample ID, does that specific sample have any other mutation also). See the image below:
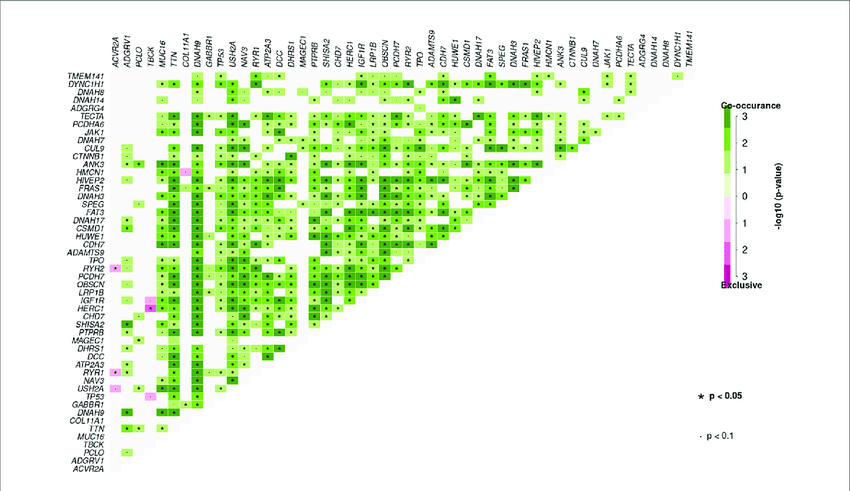
I want to get this plotted as a heatmap (example below):
I've managed to get the data into the form of the first image above, but I am completely stuck as to how to go from there to the example heatmap.
I've looked into:
df.groupby(['Sample ID', 'Protein Change', 'Cancer Type Detailed']).count().unstack('Protein Change'))
which seems to go in the right direction, but not completely there.
Basically what I want is a heatmap with Protein Change on both axis, and a count of how many times those co-exist within a single sample.
Any help would be appreciated. Thanks!!
CodePudding user response:
You could do something like this:
Sample data:
df = pd.DataFrame({'Sample ID': [1, 1, 1, 4, 4, 5, 6, 6],
'Protein Change': ['A', 'B', 'C', 'D', 'A', 'C', 'A', 'B'],
'Cancer Type Detailed': 'Some type'})
Sample ID Protein Change Cancer Type Detailed
0 1 A Some type
1 1 B Some type
2 1 C Some type
3 4 D Some type
4 4 A Some type
5 5 C Some type
6 6 A Some type
7 6 B Some type
Code:
import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
# Build co-occurrence matrix and set diagonal to zero.
ct = pd.crosstab(df['Sample ID'], df['Protein Change'])
co_occurrence = ct.T.dot(ct)
np.fill_diagonal(co_occurrence.to_numpy(), 0)
f, ax = plt.subplots(figsize=(4, 5))
# Mask lower triangular for plotting.
mask = np.tril(np.ones_like(co_occurrence))
cmap = sns.light_palette("seagreen", as_cmap=True)
sns.heatmap(co_occurrence, mask=mask, cmap=cmap, square=True, cbar_kws={"shrink": .65})
plt.show()
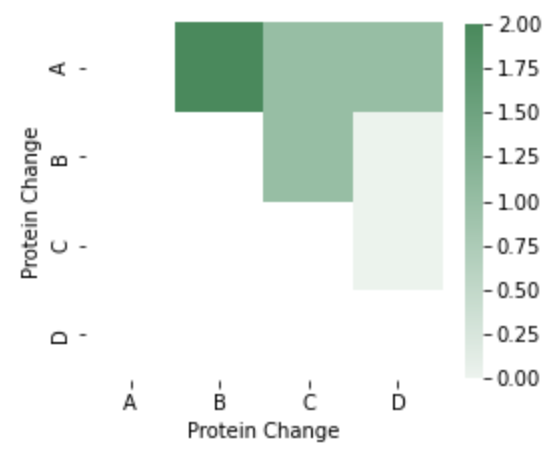
Result:
CodePudding user response:
Does this works for you?
from itertools import combinations
# create a dataframe with all combinations for each Sample ID
df_combinations = df.groupby('Sample ID').agg({'Protein Change': lambda s: list(combinations(s, 2))})
# transform the obtained dataset to a two column format
df_combinations = pd.DataFrame(df_combinations['Protein Change'].explode().dropna().tolist(), columns=['Protein Change A', 'Protein Change B'])
# count how many times each combination appears
df_counts = df_combinations.groupby(['Protein Change A', 'Protein Change B']).size().to_frame('count').reset_index()
# pivot the df to obtain the desired matrix
df_counts.pivot(index='Protein Change A', columns='Protein Change B', values='count').fillna(0)