How to make a forest plots for mixed models co-effiecents and their corresponding confidence interval. I tried this code
Model = lme (fixed = score~ Age Sex yearsofeducation walkspeed,
random = ~1|ID,
data=DB,
na.action = na.omit, method = "ML",
)
plot_summs (model)
However, I want the OR in the forest plots to be ordered in a descending fashion. Thanks for the help.
CodePudding user response:
I would call this a "coefficient plot", not a "forest plot". (A forest plot is used in meta-analyses, when you are comparing the magnitude of estimates of the same effect from many different studies.)
example setup
This is a slightly silly example, but should be close enough to yours (not clear to me why you're mentioning OR (= odds ratios?), these are typically from a logistic regression ... ?)
library(nlme)
mtcars <- transform(mtcars, cylgear = interaction(cyl, gear))
m1 <- lme(mpg ~ disp hp drat wt qsec,
random = ~1|cylgear,
data = mtcars)
coefficient plots: dotwhisker
You could get approximately what you want directly from the dotwhisker package, but it won't sort effects (or not easily, as far as I know):
library(dotwhisker)
library(broom.mixed) ## required to 'tidy' (process) lme fits
dwplot(m1, effects = "fixed")
coefficient plots: tidyverse
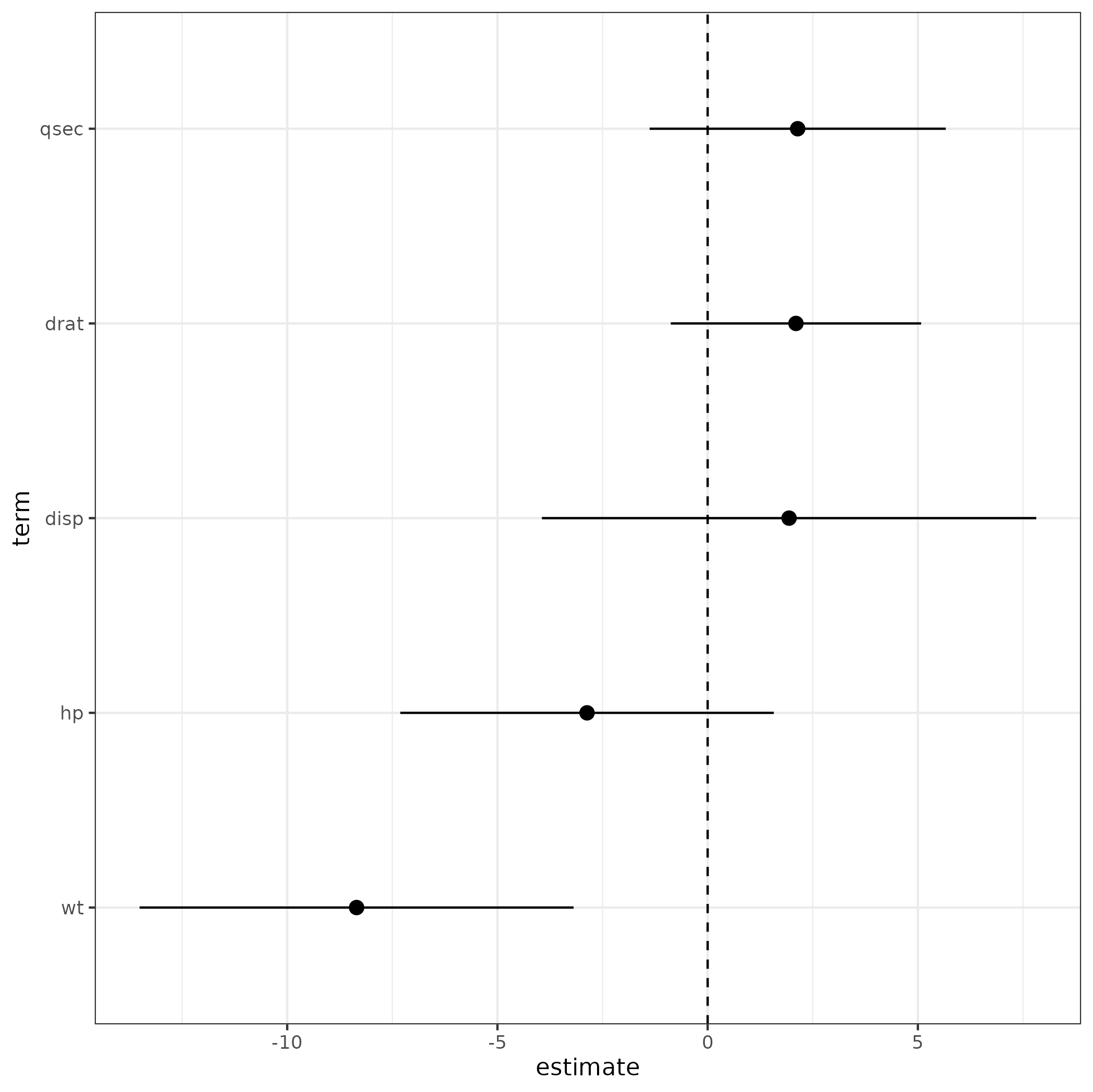
I usually do the processing myself, as I prefer increased flexibility.
library(tidyverse)
tt <- (m1
## extract estimates and CIs
|> tidy(effects = "fixed", conf.int = TRUE)
## usually *don't* want to compare intercept (dwplot does this automatically)
|> filter(term != "(Intercept)")
## scale parameters by 2SD - usually necessary for comparison
|> dotwhisker::by_2sd(data = mtcars)
## take only the bits we need, rename some (cosmetic)
|> select(term, estimate, lwr = conf.low, upr = conf.high)
## order terms by estimate value
|> mutate(across(term, ~reorder(factor(.), estimate)))
)
gg0 <- (ggplot(tt,
aes(estimate, term))
geom_pointrange(aes(xmin = lwr, xmax = upr))
geom_vline(xintercept = 0, lty = 2)
)
print(gg0)
The only remaining/possibility tricky question here is what to do if you have positive and negative coefficients of similar magnitude. If you want to sort by absolute value then
|> mutate(across(term, ~reorder(factor(.), estimate,
FUN = function(x) mean(abs(x)))
although this gets a bit ugly.
If you like the tidyverse you can substitute forcats::fct_reorder for reorder.
CodePudding user response:
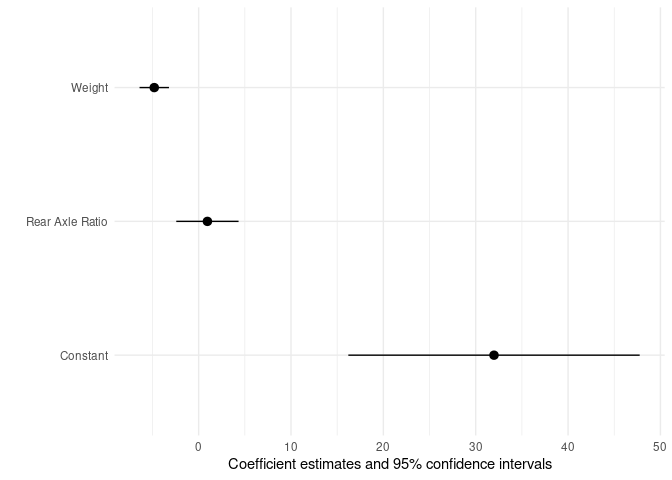
I’m just adding one more option to Ben Bolker’s excellent answer: using the modelsummary package. (Disclaimer: I am the author.)
With that package, you can use the modelplot() function to create a forest plot, and the coef_map argument to rename and reorder coefficients. If you are estimating a logit model and want the odds ratios, you can use the exponentiate argument.
The order in which you insert coefficients in the coef_map vector sorts them in the plot, from bottom to top. For example:
library(lme4)
library(modelsummary)
mod <- lmer(mpg ~ wt drat (1 | gear), data = mtcars)
modelplot(
mod,
coef_map = c("(Intercept)" = "Constant",
"drat" = "Rear Axle Ratio",
"wt" = "Weight"))