I am a R user and I have a plain text file which looks like this:
t # 0 * 3
v 0 4
v 1 7
e 0 1 1
x 0 1 2
t # 1 * 2
v 0 4
v 1 7
v 2 10
e 0 1 1
e 1 2 1
x 0 1
t # 2 * 2
v 0 7
v 1 10
e 0 1 1
x 0 2
I need to convert it into three data frames which looks like this:
# First data frame
object id number
0 0 4
0 1 7
1 0 4
1 1 7
1 2 10
2 0 7
2 1 10
# Second data frame
object from to number
0 0 1 1
1 0 1 1
1 1 2 1
2 0 1 1
# Third data frame
x t0 t1 t2
0 1 1 1
1 1 1 0
2 1 0 1
Correspondences are illustrated below:
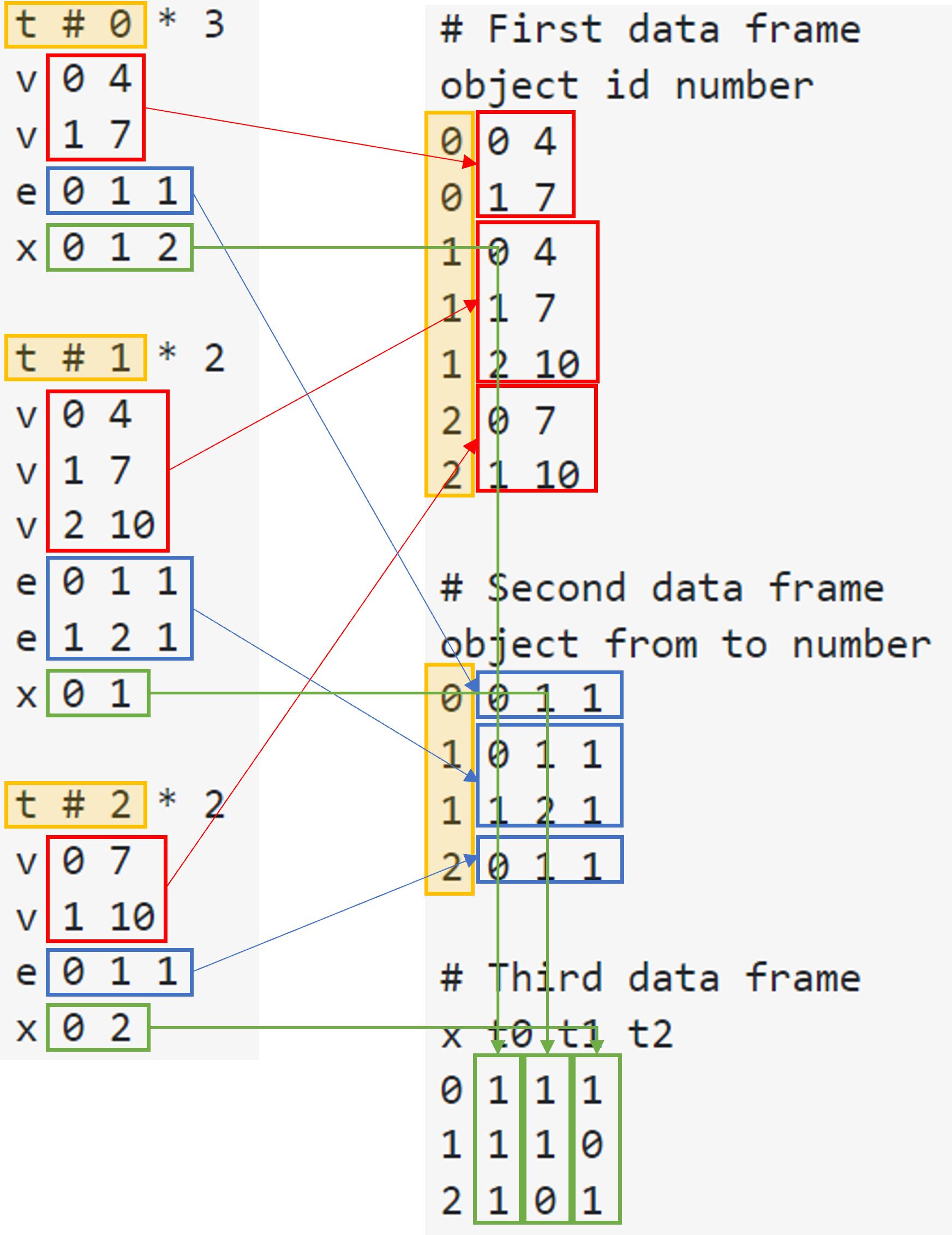
- The first data frame contains information of
vin the original file - The second data frame contains information of
ein the original file - The third data frame contains information of
xin the original file tis a identifier of eachobjectin new data frames- In the original text file, we have object
t0,t1andt2, which are indicated int1,t2andt3field in the third new data frame. In the original text file, numbers inxrow, i.e.,x 0 1 2, indicates that objectt0appears in other set of objects calledx0, 1 and 2. Set ofx(0, 1, 2) is given preliminarily.
I appreciate your suggestions to read the original text file and convert it into desired format. I could read text file using some functions such as readLines("sample.txt") and scan("sample.txt", what=character(), sep="\n", blank.lines.skip=T), however I have no knowledge to deal with these text data into data frames.
CodePudding user response:
Somehow I could make it using the following inefficient scripts. Other solutions are appreciated.
#### Load text file
result <- scan("result.txt", what=character(), sep="\n", blank.lines.skip=T)
## Line number of subgraphs
nsub <- grep("t", result)
#### Initial data frame
## Node and edge
node.s <- data.frame(matrix(ncol=3, nrow=0, dimnames=list(NULL, c("object", "id", "func"))))
edge.s <- data.frame(matrix(ncol=4, nrow=0, dimnames=list(NULL, c("object", "from", "to", "connect"))))
## Equivalence table of original graphs and subgraphs
eq.s.1 <- data.frame(t=c(0:(nrow(g)-1)))
eq.s.2 <- data.frame(matrix(ncol=length(nsub), nrow=nrow(g), dimnames=list(NULL, c(paste0("sub",c(0:(length(nsub)-1)))))))
eq.s <- cbind(eq.s.1, eq.s.2)
#### Loop to update data frames
for (i in 1:length(result)) {
## Store t number for other data frames
if (grepl("t", result[i])==1) {
t <- word(result[i],3)
}
#### Update node list (node.s)
if (grepl("v", result[i])==1) {
## Extract node
tent.v <- data.frame(object=t, id=word(result[i],2), fun=word(result[i],3))
## Append
node.s <- rbind(node.s, tent.v)
}
#### Update edge list (edge.s)
if (grepl("e", result[i])==1) {
## Extract edge
tent.e <- data.frame(object=t, from=word(result[i],2), to=word(result[i],3), connect=word(result[i],4))
## Append
edge.s <- rbind(edge.s, tent.e)
}
#### Update equivalent table (eq.s)
if (grepl("x", result[i])==1) {
## Equivalence between original graphs and subgraphs
tent.e <- data.frame(t=as.integer(unlist(strsplit(result[i], " "))[-1]), a=1)
## Update field name
names(tent.e) <- c("t",paste0("sub",t))
## Overwrite join
eq.s <- merge_dfs_overwrite_col(eq.s, tent.e, cols=names(tent.e)[2], bycol="t")
}
}
CodePudding user response:
You could do the following:
dat <- readLines('file.txt')
f <- cumsum(grepl('^t', dat))
dat1 <- paste(dat, f - 1)
df1 <- read.table(text = grep('^v', dat1, value = TRUE))[c(4,2:3)]
df1
V4 V2 V3
1 0 0 4
2 0 1 7
3 1 0 4
4 1 1 7
5 1 2 10
6 2 0 7
7 2 1 10
df2 <- read.table(text = grep('^e', dat1, value = TRUE))[c(5,2:4)]
df2
V5 V2 V3 V4
1 0 0 1 1
2 1 0 1 1
3 1 1 2 1
4 2 0 1 1
x <- read.table(text = grep('^x', dat, value = TRUE), fill=T)[-1]
df3 <- data.frame(cbind(x=0:2, apply(x, 1, \(y) 0:2 %in% y)))
df3
x V2 V3 V4
1 0 1 1 1
2 1 1 1 0
3 2 1 0 1
