Suppose I have a dataset that looks like the one below where each subject ID can have up to 4 visits (where visit=00 is baseline) and variable disease status across visits (Yes=1, No=0). This means that one subject might have one disease status at one visit but a different status at any of the follow up visits. For example, say subject ID=002 has disease (status=1) at baseline, then no disease at visits 01, 02, and then disease again at visit=03.
The issue: I need to create a graph that shows the change in disease status classification from visit to visit for each individual subject ID. But I am having some trouble visualizing how this might look like.
What I’ve done: One obvious way I was thinking of achieving this is, putting the disease status categories (yes=1, no=0) on the Y-axis and visit on X-axis and and then plot line plots for each subject ID.
Questions: I tried some SAS code and obtained the plots showed in screenshots below but not sure if this is the best way to visualize this data. And in any case, I need a way to do this in R. Also, please note that my SAS generated plot below has some issues in the sense that for ID=011 for example, the visit numbers on the x-axis are not in order, i.e. they should be showing up as 00, 01, 02, 03 instead of 00, 02, 03, 01.
Any help with generating a plot in R for this would be much appreciated.
Thanks!
ID<-c(001, 001, 001, 001, 002, 002, 002, 002, 003, 003, 004, 004, 004, 005, 005, 005, 005, 006, 007, 007, 008, 008, 008, 009, 009, 009, 009, 010, 011, 011)
Visit<-c(00, 01, 02, 03, 00, 01, 02, 03, 01, 02, 00, 02, 03, 00, 01, 02, 03, 02, 00, 01, 00, 02, 03, 00, 01, 02, 03, 00, 01, 03)
Status<- c(0, 0, 0, 0, 1, 0, 0, 1, 1, 1, 1, 1, 0, 1, 0, 0, 0, 0, 0, 0, 1, 1, 0, 0, 1, 0, 1, 1, 0, 1)
data<-data.frame(ID,Visit, Status)
View(data)
CodePudding user response:
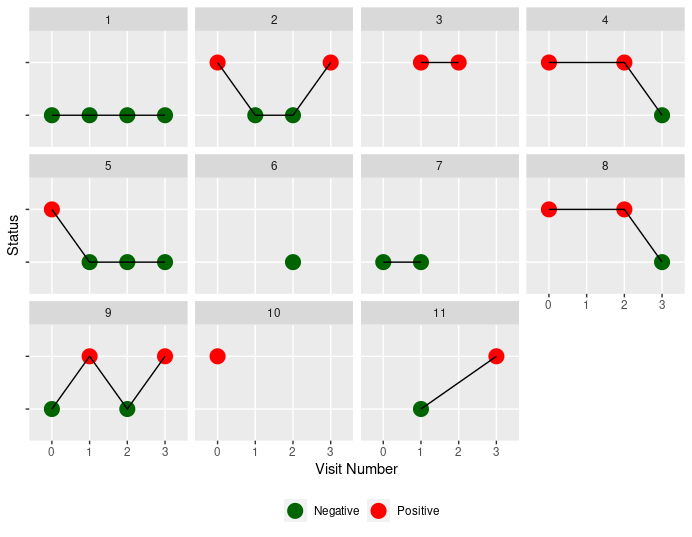
Here is one way to visualize:
ggplot(data, aes(as.factor(Visit), as.factor(Status), group=ID))
geom_point(size=5, aes(color=ifelse(Status==1, "Positive", "Negative")))
geom_line()
scale_color_manual(values=c("Negative"="darkgreen", "Positive"="red"))
facet_wrap(~ID)
scale_x_discrete(breaks=c(0,1,2,3))
labs(x="Visit Number", y="Status", color="")
theme(legend.position = "bottom", axis.text.y = element_blank())
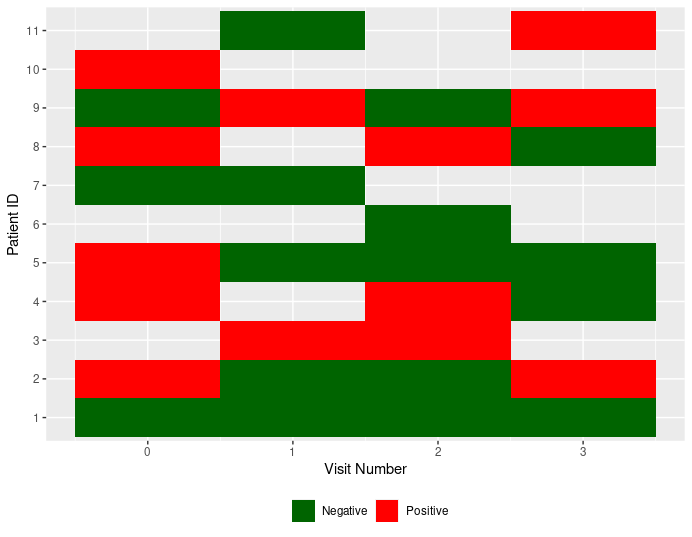
Another option is like this:
ggplot(data, aes(
Visit, as.factor(ID), group=ID,
fill=ifelse(Status==1, "Positive", "Negative")))
geom_tile()
scale_fill_manual(values=c("Negative"="darkgreen", "Positive"="red"))
labs(x="Visit Number", y="Patient ID", fill="")
theme(legend.position = "bottom")