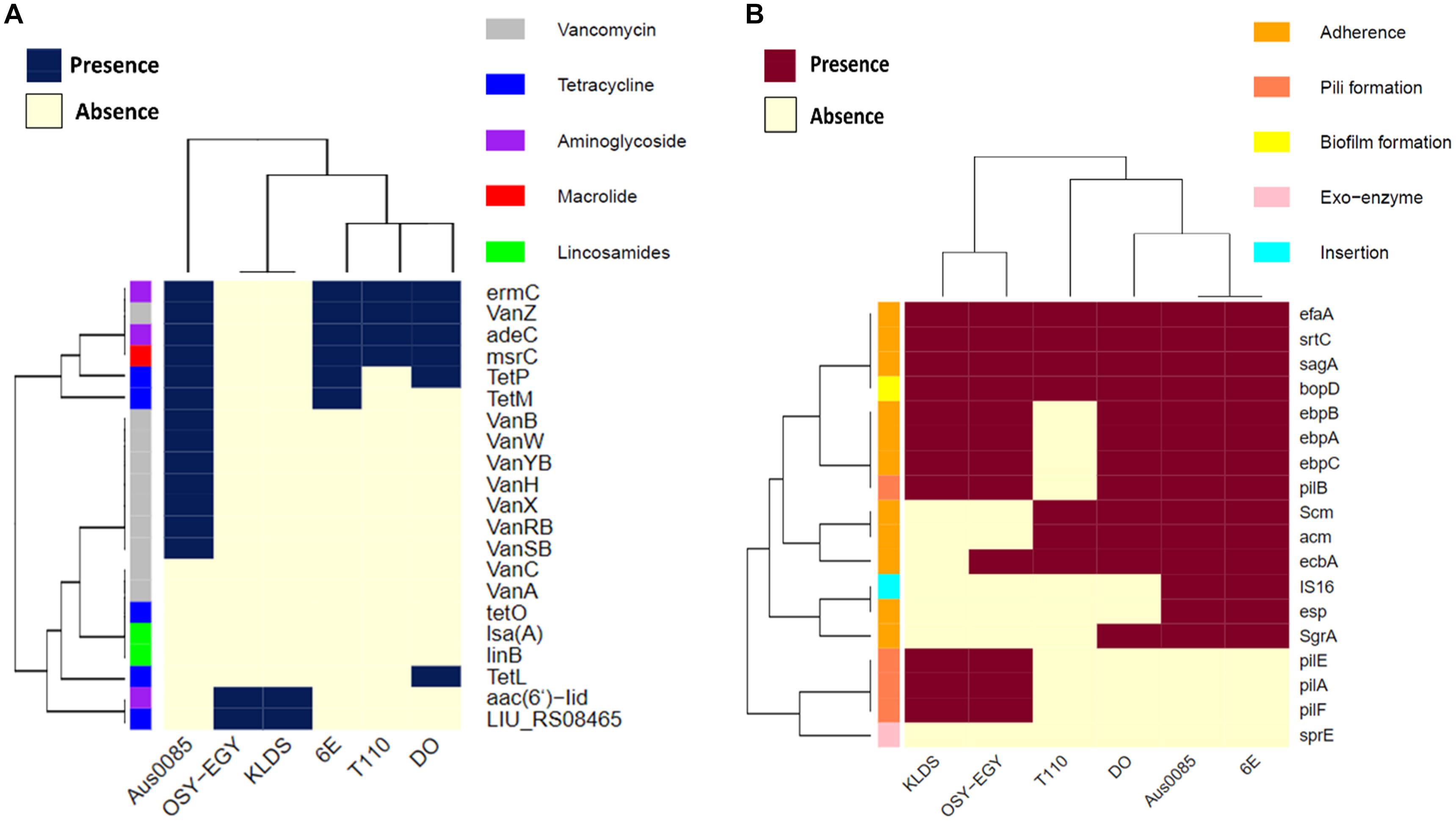
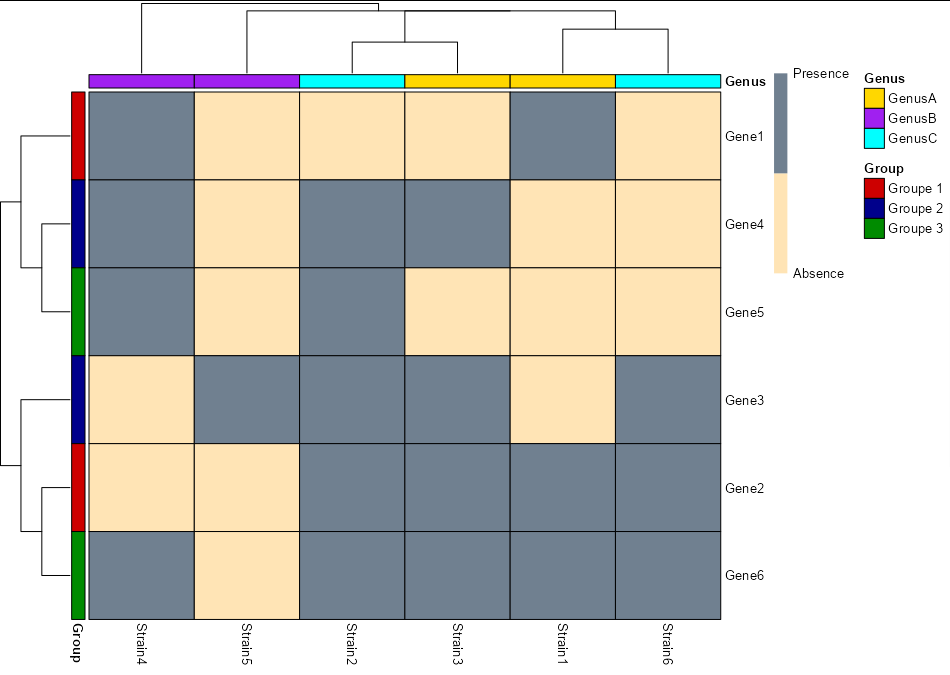
I need help, I'm new to R and have built a heatmap but I can't seem to set up groups next to the rows and columns. Please I want something like this heatmap.
#Design of the data as an example
Data <- data.frame(replicate(6,sample(0:1,6,rep=TRUE)))
row.names(Data) <- c('Gene1','Gene2','Gene3','Gene4','Gene5','Gene6')
colnames(Data) <- c("Strain1", "Strain2", "Strain3", "Strain4", "Strain5", "Strain6")
pheatmap(Data, color=colorRampPalette(c("#FFE4B5","#708090"))(2),
legend_breaks = c(1, 0),
legend_labels = c("Presence", "Absence"),
border_color = "black", display_numbers = FALSE,
number_color = "black",
fontsize_number = 8)
#Now I want to create Groups (Row) ##To designate that they are
part of the same antibiotic resistance family##
#Groupe1 "Gene3 and Gene6" Color1
#Groupe2 "Gene1 and Gene5" Color2
#Groupe3 "Gene2 and Gene4" Color3
#How can I do this?
#Same question if I want to create Groupes for column ##To
designate that they are part of the same bacterial genus#
#Band1 "Strain1" and "Strain3" Color4
#Band2 "Strain4" and "Strain5" Color5
#Band3 "Strain2" and "Strain6" Color6
CodePudding user response:
You need to create little data frames for your annotations and pass them to annotation_row and annotation_col. The actual colours are passed as a named list of two named vectors to annotation_colors:
row_df <- data.frame(Group = paste("Groupe", rep(1:3, each = 2)),
row.names = paste0("Gene", 1:6))
col_df <- data.frame(Genus = c("GenusA", "GenusC", "GenusA",
"GenusB", "GenusB", "GenusC"),
row.names = paste0("Strain", 1:6))
pheatmap(Data,
color = colorRampPalette(c("#FFE4B5","#708090"))(2),
legend_breaks = c(1, 0),
legend_labels = c("Presence", "Absence"),
border_color = "black", display_numbers = FALSE,
number_color = "black",
annotation_row = row_df,
annotation_col = col_df,
fontsize_number = 8,
annotation_colors = list(Group = c("Groupe 1" = "red3", "Groupe 2" = "blue4",
"Groupe 3" = "green4"),
Genus = c(GenusA = "gold", GenusB = "purple",
GenusC = "cyan")))