I have created a GAM and set up the predictions but having trouble with how to plot any smooth functions from my model. Been trying to plot these in ggplot but having trouble with the arguments/aesthetics now I have added in a the month aswell, seening some people say to use geom_smooth() too but I'm not sure. If anyone can advise me on this that would be great, i had added my data, model and predictions below;
model
mod = gam(co2 ~ s(timeStep, k = 200, bs = "cs") s(month, k = 12, bs = "cc"),
data = carbonD,
family = gaussian(link = "identity"))
predictions
#create predictions
preds = predict(mod, type = 'terms', se.fit = TRUE)
#combine our predictions with coefficients
fit = preds$fit coef(mod)[1]
data snipet
carbonD
co2 month year timeStep
1 315.42 1 1959 1
2 316.31 2 1959 2
3 316.50 3 1959 3
4 317.56 4 1959 4
5 318.13 5 1959 5
6 318.00 6 1959 6
7 316.39 7 1959 7
8 314.65 8 1959 8
9 313.68 9 1959 9
10 313.18 10 1959 10
11 314.66 11 1959 11
12 315.43 12 1959 12
13 316.27 1 1960 13
14 316.81 2 1960 14
15 317.42 3 1960 15
CodePudding user response:
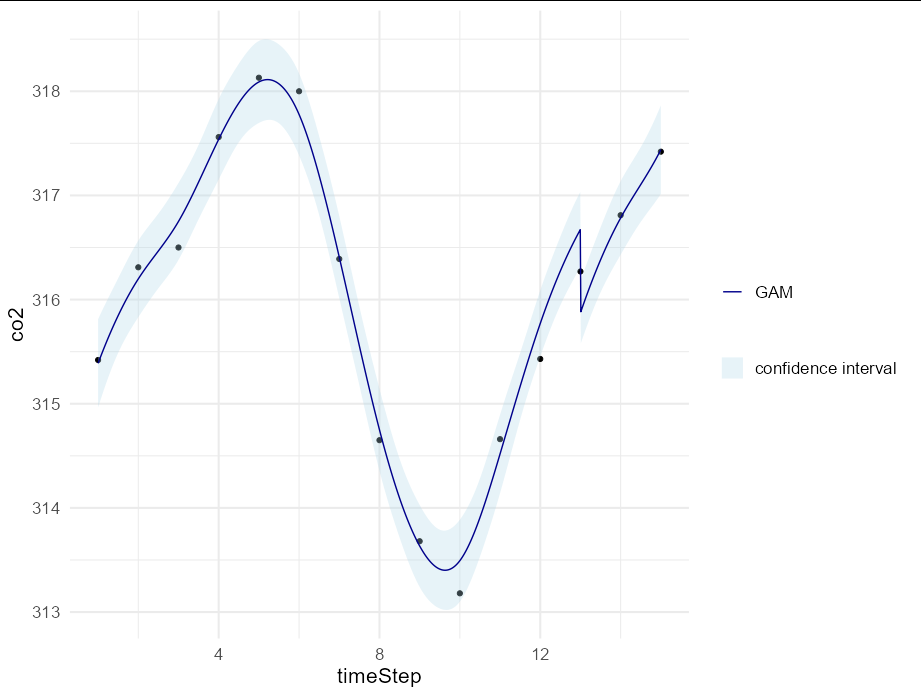
There are two ways to plot your exact model in ggplot. One is to use geom_smooth, but you can't do this with two variables on the right hand side. Actually, in your case it would be possible because month is calculable from the time step, but let's ignore that for now and just plot your model predictions directly using a ribbon and a line.
First, load the required packages and create the model (note because we only have a snippet of your data, I have had to reduce the number of knots)
library(mgcv)
library(ggplot2)
mod = gam(co2 ~ s(timeStep, k = 4, bs = "cs") s(month, k = 12, bs = "cc"),
data = carbonD,
family = gaussian(link = "identity"))
Now we create a little data frame of the values we want our predictions at, with 1000 points across the range of our data:
newdata <- data.frame(timeStep = seq(1, 15, length.out = 1000),
month = (seq(1, 15, length.out = 1000) - 1) %% 12 1)
Now we make our predictions and use the standard error fit to create an upper and lower confidence band.
pred <- predict(mod, newdata, type = 'response', se.fit = TRUE)
newdata$co2 <- pred$fit
newdata$lower <- pred$fit - 1.96 * pred$se.fit
newdata$upper <- pred$fit 1.96 * pred$se.fit
Now we can plot our results:
ggplot(carbonD, aes(timeStep, co2))
geom_point()
geom_ribbon(data = newdata, alpha = 0.3,
aes(ymin = lower, ymax = upper, fill = "confidence interval"))
geom_line(data = newdata, aes(color = "GAM"))
scale_fill_manual(values = "lightblue", name = NULL)
scale_color_manual(values = "darkblue", name = NULL)
theme_minimal(base_size = 16)
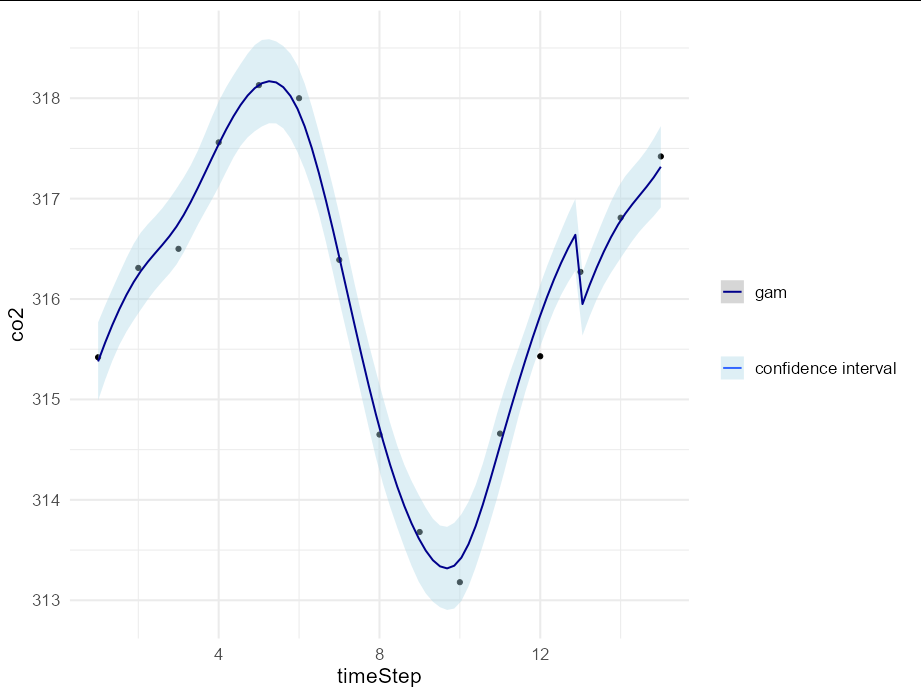
It is also possible to use your gam within geom_smooth directly, but you need to be able to express the model in terms of y and x, where x is the time step. You can get the month by subtracting 1 from the time step, getting this number modulo 12, and adding 1 again, so it is possible to avoid explicitly creating a prediction data frame, at the cost of making the plotting code more complex:
ggplot(carbonD, aes(timeStep, co2))
geom_point()
geom_smooth(formula = y ~ s(x, k = 4, bs = "cs")
s((x - 1) %% 12 1, k = 12, bs = "cc"),
method = "gam", size = 0.7,
method.args = list(family = gaussian(link = "identity")),
aes(color = "gam", fill = "confidence interval"))
scale_fill_manual(values = "lightblue", name = NULL)
scale_color_manual(values = "darkblue", name = NULL)
theme_minimal(base_size = 16)
As a caveat to this, it is not clear to me that you should have both month and timestep, since one is just the modulus of the other. It may be better to use just timestep alone, or use year and month if you want to separate the long-term and seasonal effects.
CodePudding user response:
The simplest way would be to use geom_smooth with LOESS: geom_smooth(method="loess", span=0.5) and play with the span parameter to get a more smooth or wiggly shape.