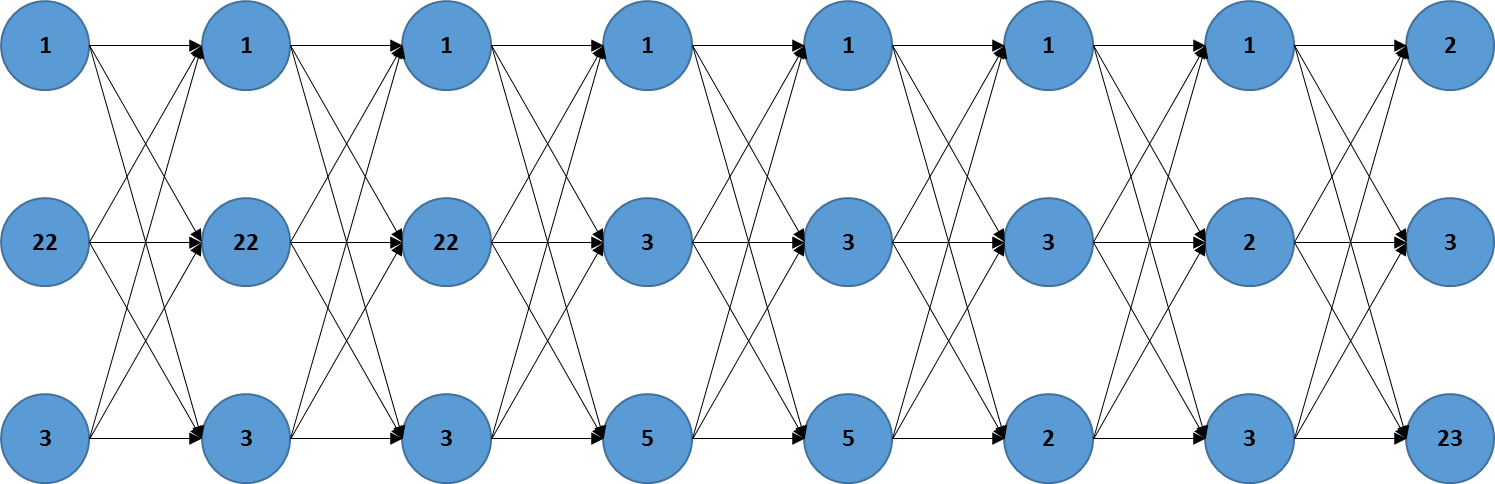
I want to create a directed adjacency matrix from data like this:
| x1 | x2 | x3 | x4 | x5 | x6 | x7 | x8 |
|---|---|---|---|---|---|---|---|
| 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 |
| 22 | 22 | 22 | 3 | 3 | 3 | 2 | 3 |
| 3 | 3 | 3 | 5 | 5 | 2 | 3 | 23 |
Where the columns represent states in time.
The adjacency matrix should reflect the following logic:
For the column x1: 1 should go to the 3 rows in column x2,
22 should go to the 3 rows in column x2,
3 should go to the 3 rows in column x2
For the column x2: The same pattern going to column x3. And this for all columns. So it's like linking each element in a given column to all elements of the following column, and so on.
The output should be a matrix with columns and rows N x N (where N in the number of unique values in the whole matrix) and... well, an adjacency matrix.
This dataframe is just a sample, the one I have to use has hundreds of columns.
For these 8 columns, the output should resemble something like this:
| 1 | 2 | 3 | 5 | 22 | 23 | |
|---|---|---|---|---|---|---|
| 1 | 6 | 1 | 0 | 0 | 0 | 0 |
| 2 | 0 | 0 | 2 | 0 | 0 | 0 |
| 3 | 0 | 1 | 4 | 1 | 0 | 1 |
| 5 | 0 | 1 | 0 | 1 | 0 | 0 |
| 22 | 0 | 0 | 1 | 0 | 2 | 0 |
| 23 | 0 | 0 | 0 | 0 | 0 | 0 |
This is a representation of how the graph should look like. (edited)
I've been trying to make it work, but am really lost by now... TIA
P.S. I'm working with R but Python could also work.
CodePudding user response:
It seems that you may misunderstand how an adjacency matrix works.
The matrix contains Boolean values ( true or false )
The nodes should be indexed 1,2,3,4, ...
If there is a link from node 1 to node 2, then the cell in row 2, column 1 will be true.
Let's index your first two columns like this
1 4
2 5
3 6
So node 1 is linked to nodes 4,5, and 6
and the adjacency matrix looks like this
1 2 3 4 5 6
1
2
3
4 1 1 1
5 1 1 1
6 1 1 1
CodePudding user response:
I don't think the adjacency matrix is the thing you are after. I guess it should be the summary info of transitions. You can try the base R code below (without igraph)
d <- do.call(
rbind,
apply(
embed(seq_along(df), 2),
1,
function(k) {
expand.grid(
setNames(
df[rev(k)],
c("from", "to")
)
)
}
)
)
lvls <- sort(unique(unlist(d)))
table(list2DF(lapply(d, factor, level = lvls)))
which gives
to
from 1 2 3 5 22 23
1 6 3 7 2 2 1
2 1 2 2 0 0 1
3 6 3 7 2 2 1
5 2 1 2 1 0 0
22 3 0 3 1 2 0
23 0 0 0 0 0 0
data
> dput(df)
structure(list(x1 = c(1L, 22L, 3L), x2 = c(1L, 22L, 3L), x3 = c(1L,
22L, 3L), x4 = c(1L, 3L, 5L), x5 = c(1L, 3L, 5L), x6 = c(1L,
3L, 2L), x7 = 1:3, x8 = c(2L, 3L, 23L)), class = "data.frame", row.names = c(NA,
-3L))
CodePudding user response:
You could do:
as.data.frame.matrix(xtabs(~factor(x1, unique(c(x1, values))) values, cbind(df[1], stack(df[-1]))))
1 2 3 5 22 23
1 6 1 0 0 0 0
22 0 1 4 0 2 0
3 0 1 3 2 0 1
5 0 0 0 0 0 0
2 0 0 0 0 0 0
23 0 0 0 0 0 0
xtabs(~x1 x, transform(reshape(df, names(df)[-1], dir='long', sep=''), x1 = factor(x1, unique(c(x,x1)))))
x
x1 1 2 3 5 22 23
1 6 1 0 0 0 0
22 0 1 4 0 2 0
3 0 1 3 2 0 1
5 0 0 0 0 0 0
2 0 0 0 0 0 0
23 0 0 0 0 0 0
library(tidyverse)
df %>%
mutate(x1 = factor(x1, unique(unlist(.)))) %>%
pivot_longer(-x1) %>%
xtabs(~x1 value,.) %>%
as.data.frame.matrix()
1 2 3 5 22 23
1 6 1 0 0 0 0
22 0 1 4 0 2 0
3 0 1 3 2 0 1
5 0 0 0 0 0 0
2 0 0 0 0 0 0
23 0 0 0 0 0 0