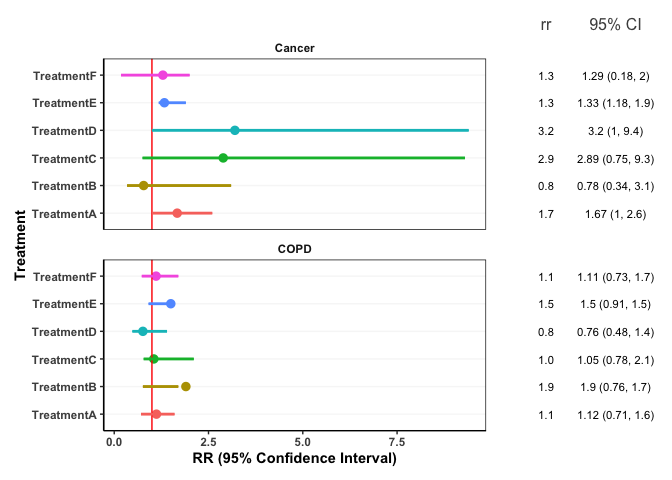
I am new to R and have created a forest/interval plot and am including a table beside the plot with my confidence intervals and risk ratios. My issue is that the RR and CIs do not line up exactly with the horizontal grid lines on the plot. I have tried to use the patchwork solution as seen here, but that doesn't seem to work for me: 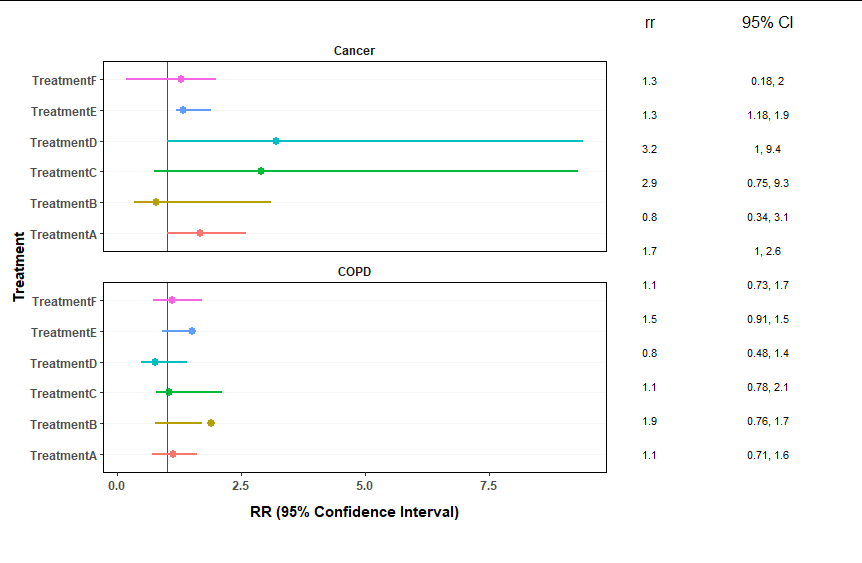
CodePudding user response:
The main issue IMHO is that you made your table columns as two separate plots. Instead one option would be to make you table plot as one plot and importantly to facet by X too in the table plot. Otherwise the table plot is lacking the strip texts and without IMHO it's nearly impossible to align the table rows with the point ranges. The rest is styling where it's important to not get simply rid of theme elements, e.g. for the alignment it's important that there are strip boxes so we can't use element_blank but instead have to use empty strings for the strip texts.
tester <- data.frame(
treatmentgroup = c("TreatmentA", "TreatmentB", "TreatmentC", "TreatmentD", "TreatmentE", "TreatmentF", "TreatmentA", "TreatmentB", "TreatmentC", "TreatmentD", "TreatmentE", "TreatmentF"),
rr = c(1.12, 1.9, 1.05, 0.76, 1.5, 1.11, 1.67, 0.78, 2.89, 3.2, 1.33, 1.29),
low_ci = c(0.71, 0.76, 0.78, 0.48, 0.91, 0.73, 1, 0.34, 0.75, 1, 1.18, 0.18),
up_ci = c(1.6, 1.7, 2.11, 1.4, 1.5, 1.7, 2.6, 3.1, 9.3, 9.4, 1.9, 2),
RR_ci = c(
"1.12 (0.71, 1.6)", "1.9 (0.76, 1.7)", "1.05 (0.78, 2.1)", "0.76 (0.48, 1.4)", "1.5 (0.91, 1.5)", "1.11 (0.73, 1.7)",
"1.67 (1, 2.6)", "0.78 (0.34, 3.1)", "2.89 (0.75, 9.3)", "3.2 (1, 9.4)", "1.33 (1.18, 1.9)", "1.29 (0.18, 2)"
),
ci = c(
"0.71, 1.6",
"0.76, 1.7",
"0.78, 2.1",
"0.48, 1.4",
"0.91, 1.5",
"0.73, 1.7",
"1, 2.6",
"0.34, 3.1",
"0.75, 9.3",
"1, 9.4",
"1.18, 1.9",
"0.18, 2"
),
X = c("COPD", "COPD", "COPD", "COPD", "COPD", "COPD", "Cancer", "Cancer", "Cancer", "Cancer", "Cancer", "Cancer"),
no = c(1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12)
)
# Reduce the opacity of the grid lines: Default is 255
col_grid <- rgb(235, 235, 235, 100, maxColorValue = 255)
library(dplyr, warn = FALSE)
library(ggplot2)
library(patchwork)
forest <- ggplot(
data = tester,
aes(x = treatmentgroup, y = rr, ymin = low_ci, ymax = up_ci)
)
geom_pointrange(aes(col = treatmentgroup))
geom_hline(yintercept = 1, colour = "red")
xlab("Treatment")
ylab("RR (95% Confidence Interval)")
geom_errorbar(aes(ymin = low_ci, ymax = up_ci, col = treatmentgroup), width = 0, cex = 1)
facet_wrap(~X, strip.position = "top", nrow = 9, scales = "free_y")
theme_classic()
theme(
panel.background = element_blank(), strip.background = element_rect(colour = NA, fill = NA),
strip.text.y = element_text(face = "bold", size = 12),
panel.grid.major.y = element_line(colour = col_grid, size = 0.5),
strip.text = element_text(face = "bold"),
panel.border = element_rect(fill = NA, color = "black"),
legend.position = "none",
axis.text = element_text(face = "bold"),
axis.title = element_text(face = "bold"),
plot.title = element_text(face = "bold", hjust = 0.5, size = 13)
)
coord_flip()
dat_table <- tester %>%
select(treatmentgroup, X, RR_ci, rr) %>%
mutate(rr = sprintf("%0.1f", round(rr, digits = 1))) %>%
tidyr::pivot_longer(c(rr, RR_ci), names_to = "stat") %>%
mutate(stat = factor(stat, levels = c("rr", "RR_ci")))
table_base <- ggplot(dat_table, aes(stat, treatmentgroup, label = value))
geom_text(size = 3)
scale_x_discrete(position = "top", labels = c("rr", "95% CI"))
facet_wrap(~X, strip.position = "top", ncol = 1, scales = "free_y", labeller = labeller(X = c(Cancer = "", COPD = "")))
labs(y = NULL, x = NULL)
theme_classic()
theme(
strip.background = element_blank(),
panel.grid.major = element_blank(),
panel.border = element_blank(),
axis.line = element_blank(),
axis.text.y = element_blank(),
axis.text.x = element_text(size = 12),
axis.ticks = element_blank(),
axis.title = element_text(face = "bold"),
)
forest table_base plot_layout(widths = c(10, 4))