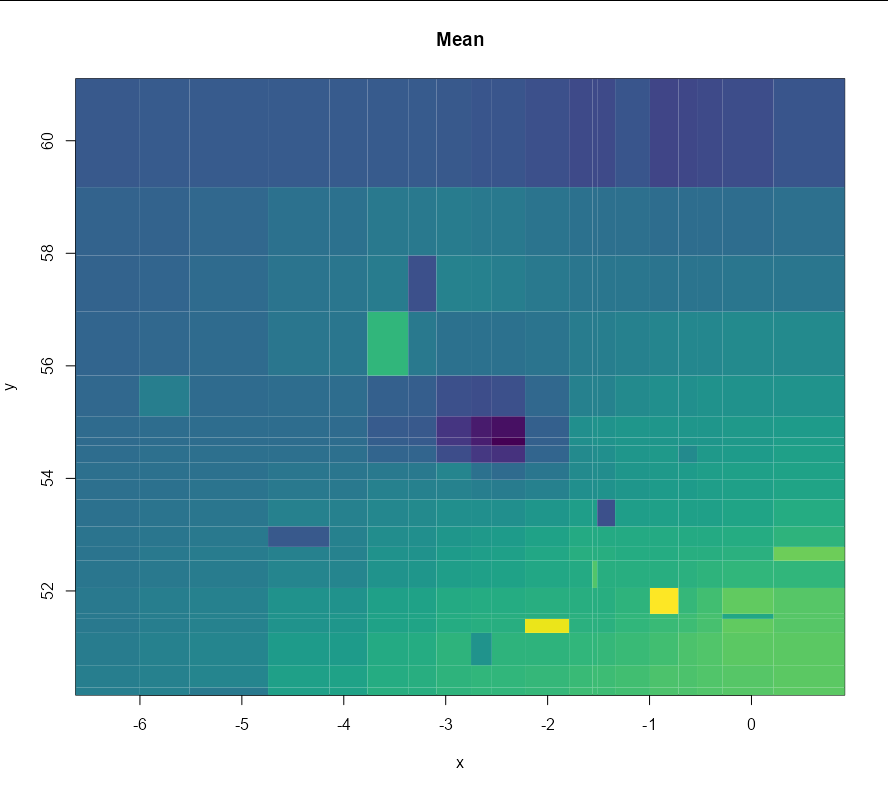
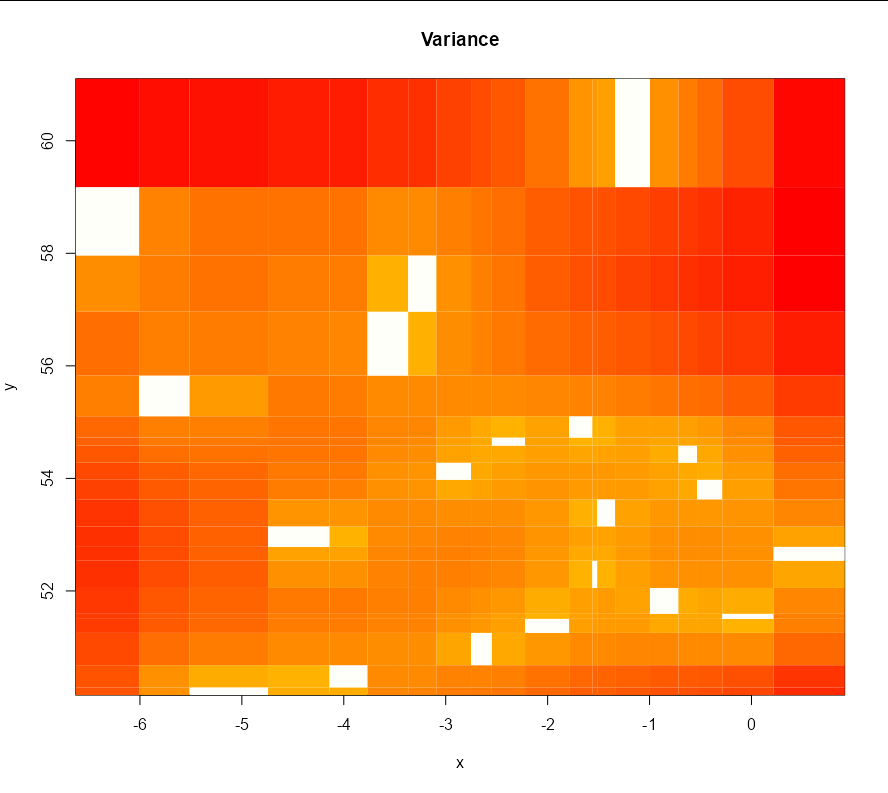
I have created the following model and predictions but I'm having trouble with the code to plot the predictions. I think it's a dimensions issue, does anyone know the changes I need to make for this to work?
code used;
#variogram
summer_vario = variog(geo_summer_df2, option = 'bin', estimator.type='modulus', bin.cloud = TRUE)
#fitting a basic parametric model
defult_summer_mod = variofit(summer_vario)
#creating predictions
preds_grid = matrix(c(-5.697, 55.441, -0.807, 51.682, -5.328, 50.218, -2.451, 54.684, -4.121, 50.355, -1.586, 54.768, -0.131, 51.505, -4.158, 52.915,
-0.442, 53.875, -3.413, 56.214, -2.860, 54.076, -3.323, 57.711, 0.566, 52.651, -0.626, 54.481, -1.185, 60.139, -2.643, 51.006,
-1.491, 53.381, -1.536, 52.424, -6.319, 58.213, -1.992, 51.503), nrow = 20, byrow = TRUE)
summer_preds = krige.conv(geo_summer_df2, locations = preds_grid, krige = krige.control(obj.model = defult_summer_mod))
#plotting predictions
#mean
image(summer_preds, col = viridis::viridis(100), zlim = c(100, max(c(summer_preds$predict))),
coords.data = geo_summer_df2[1]$coords, main = 'Mean', xlab = 'x', ylab = 'y',
x.leg = c(700, 900), y.leg = c(20, 70))
#variation
image(summer_preds, values = summer_preds$krige.var, col = heat.colors(100)[100:1],
zlim = c(0,max(c(summer_preds$krige.var))), coords.data = geo_summer_df2[1]$coords,
main = 'Variance', xlab = 'x', ylab = 'y', x.leg = c(700, 900), y.leg = c(20, 70))
data used;
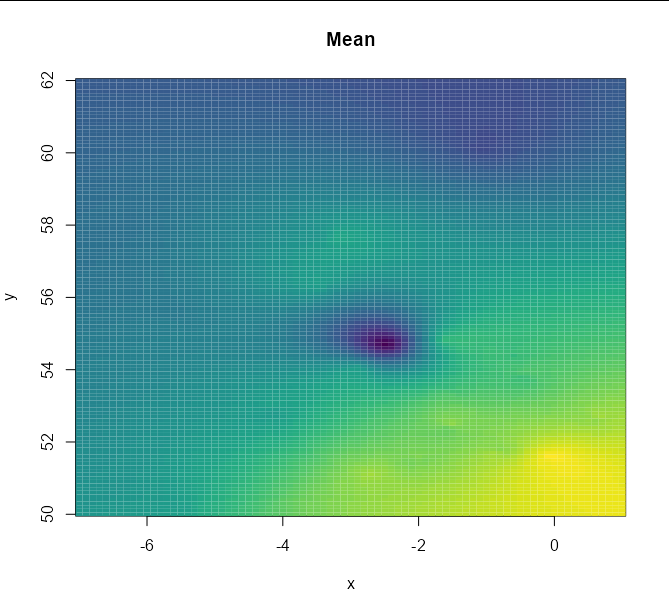
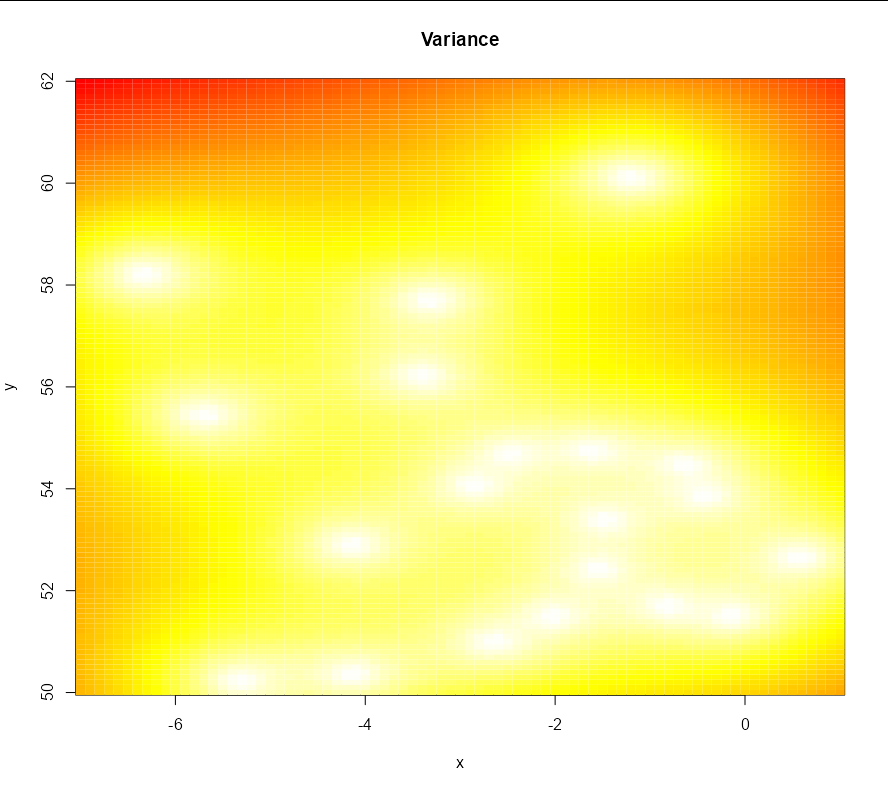
image(xvals, yvals, matrix(summer_preds$krige.var, nrow = length(xvals)),
col = heat.colors(100)[100:1], main = 'Variance', xlab = 'x', ylab = 'y')
Note that you will get better images if you use a finely-spaced sequence for x and y:
xvals <- seq(-7, 1, 0.1)
yvals <- seq(50, 62, 0.1)
The plots this produces with the same code otherwise are: