I have the following data:
structure(list(cells = c("Adipocytes", "B-cells", "Basophils",
"CD4 memory T-cells", "CD4 naive T-cells", "CD4 T-cells",
"CD4 Tcm", "CD4 Tem", "CD8 naive T-cells", "CD8 T-cells",
"CD8 Tcm", "Class-switched memory B-cells", "DC", "Endothelial cells",
"Eosinophils", "Epithelial cells", "Fibroblasts", "Hepatocytes",
"ly Endothelial cells", "Macrophages", "Macrophages M1", "Macrophages M2",
"Mast cells", "Melanocytes", "Memory B-cells", "Monocytes", "mv Endothelial cells",
"naive B-cells", "Neutrophils", "NK cells", "pDC", "Pericytes",
"Plasma cells", "pro B-cells", "Tgd cells", "Th1 cells", "Th2 cells",
"Tregs"), Response = c(0, 8, 0, 5, 4, 4, 3, 3, 2, 3, 8, 5, 3,
0, 1, 1, 0, 2, 1, 5, 3, 3, 2, 2, 7, 4, 3, 5, 2, 2, 8, 0, 1, 2,
3, 3, 2, 8), No_Response = c(6, 0, 1, 1, 2, 2, 1, 3, 3, 1, 1,
2, 1, 2, 3, 5, 2, 2, 3, 1, 1, 2, 2, 1, 0, 0, 5, 0, 1, 0, 0, 3,
1, 1, 5, 3, 1, 3)), class = "data.frame", row.names = c(NA, -38L
))
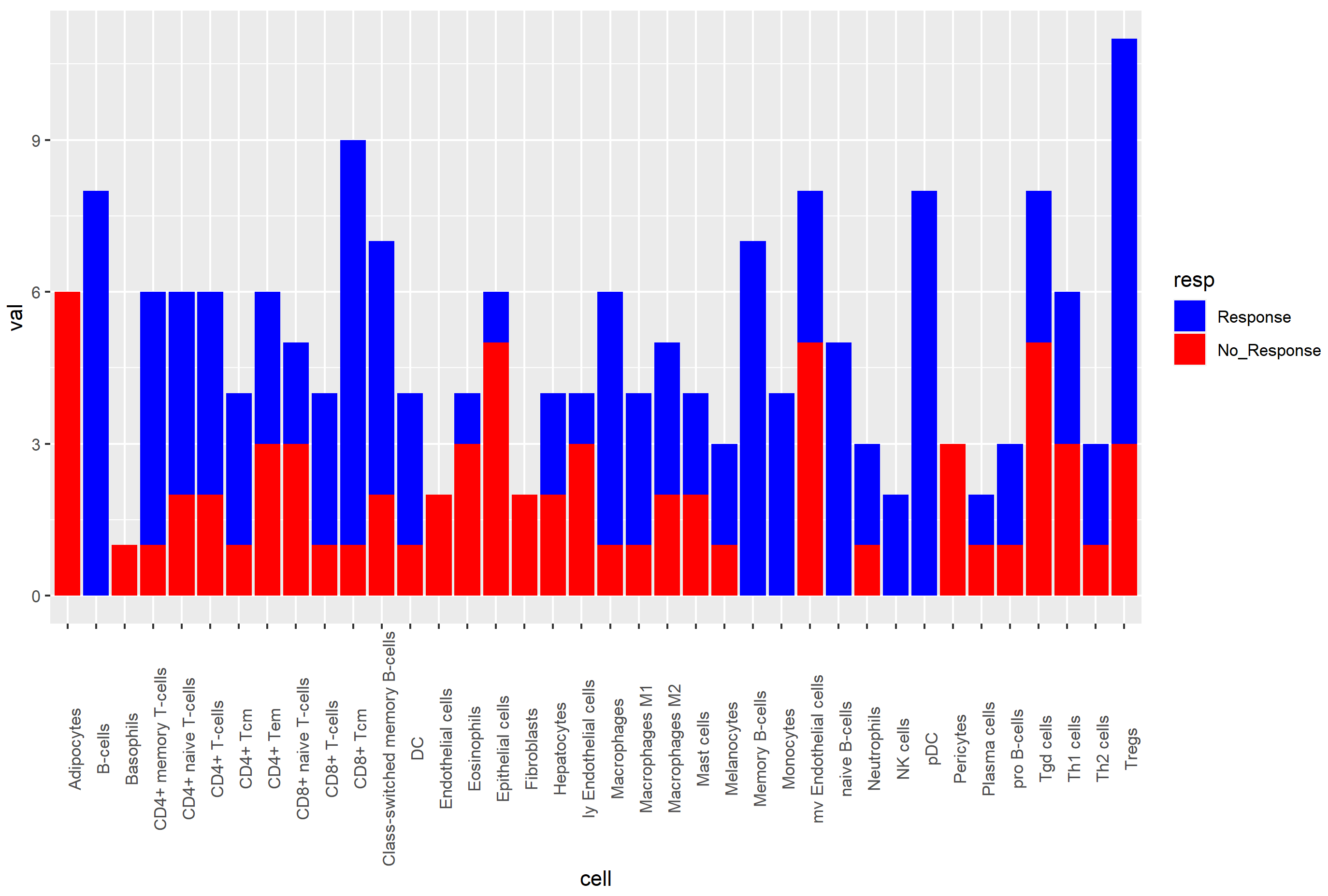
I want to make a bar chart, so that for each cell type, I get the Response number in blue, and the No_Response number in red. Something that looks like this more or less: (cells in the x-axis and the values in the y-axis):
CodePudding user response:
library(ggplot2)
data <-
structure(
list(
cells = c(
"Adipocytes",
"B-cells",
"Basophils",
"CD4 memory T-cells",
"CD4 naive T-cells",
"CD4 T-cells",
"CD4 Tcm",
"CD4 Tem",
"CD8 naive T-cells",
"CD8 T-cells",
"CD8 Tcm",
"Class-switched memory B-cells",
"DC",
"Endothelial cells",
"Eosinophils",
"Epithelial cells",
"Fibroblasts",
"Hepatocytes",
"ly Endothelial cells",
"Macrophages",
"Macrophages M1",
"Macrophages M2",
"Mast cells",
"Melanocytes",
"Memory B-cells",
"Monocytes",
"mv Endothelial cells",
"naive B-cells",
"Neutrophils",
"NK cells",
"pDC",
"Pericytes",
"Plasma cells",
"pro B-cells",
"Tgd cells",
"Th1 cells",
"Th2 cells",
"Tregs"
),
Response = c(
0,
8,
0,
5,
4,
4,
3,
3,
2,
3,
8,
5,
3,
0,
1,
1,
0,
2,
1,
5,
3,
3,
2,
2,
7,
4,
3,
5,
2,
2,
8,
0,
1,
2,
3,
3,
2,
8
),
No_Response = c(
6,
0,
1,
1,
2,
2,
1,
3,
3,
1,
1,
2,
1,
2,
3,
5,
2,
2,
3,
1,
1,
2,
2,
1,
0,
0,
5,
0,
1,
0,
0,
3,
1,
1,
5,
3,
1,
3
)
),
class = "data.frame",
row.names = c(NA,-38L)
)
df <- cbind(data[, 1], data[, 2], rep("Response", nrow(data)))
df <-
as.data.frame(rbind(df, cbind(data[, 1], data[, 3], rep(
"No_Response", nrow(data)
))))
colnames(df) <- c("cells", "quant.response", "type.response")
p <-
ggplot(data = df, aes(x = cells, y = quant.response, fill = type.response))
geom_bar(stat = "identity")
print(p)
CodePudding user response:
library(ggplot2)
library(data.table)
setDT(df)
df <- melt(df, id.vars = c("cells"), measure.vars = c("Response", "No_Response"))
colnames(df) <- c("cell", "resp", "val")
ggplot(df, aes(x = cell, y = val, fill = resp))
geom_bar(stat = "identity")
scale_fill_manual(values = c("blue", "red"))
theme(axis.text.x = element_text(angle = 90))