I have 2 pandas dataframes, df1 and df2 which both have data from 2 different days between 21:00 and 8:00. The data should be 1 data point per minute, however there are there are missing values e.g.
location time Data
0 1 21:00:00 8
1 1 21:02:00 6
the data point for 21:01:00 does not exist.
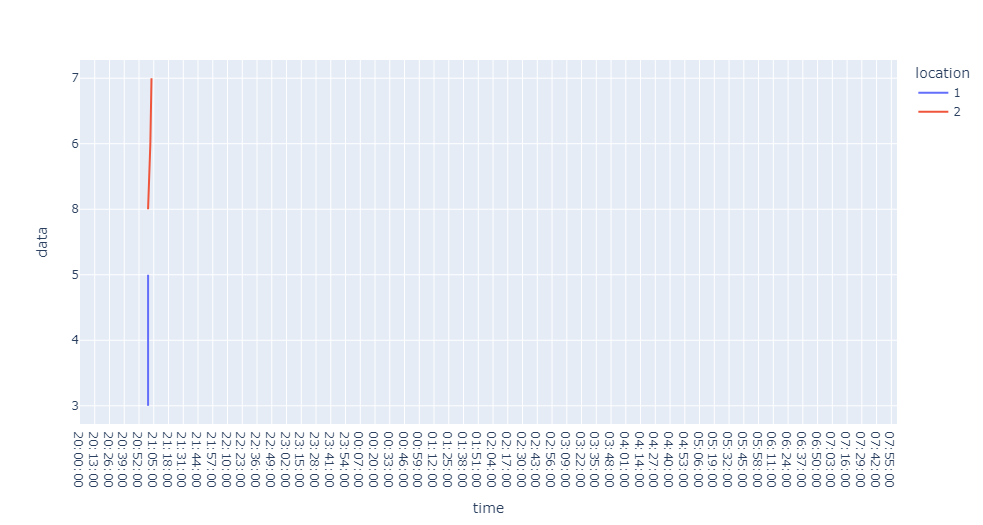
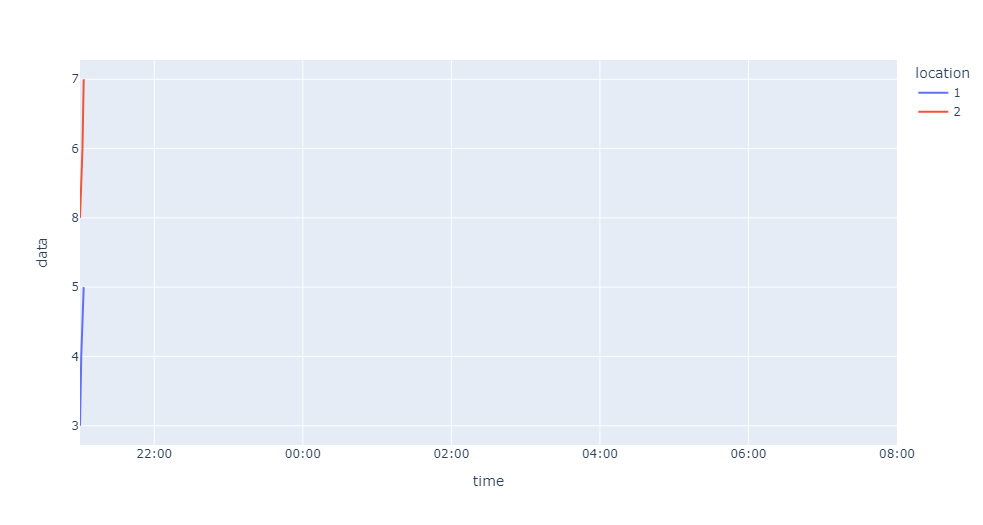
The missing data points occur at different times for each of the dataframes, so when I try to plot both of them on the same plot this happens:
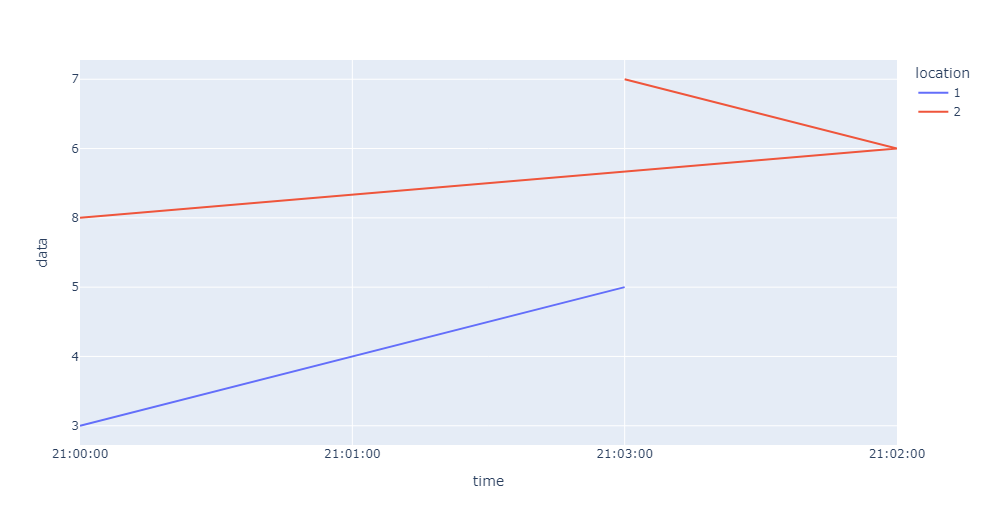
If I plot them individually they're both correct. I think the horizontal red lines are caused by the time values that exist in the red dataframe but not in the blue dataframe.
Has anyone encountered this before? I want to plot both of them on the same axis, starting at 21:00 and finishing at 08:00.
Here is the code I'm using:
import pandas as pd
import plotly.express as px
df1 = pd.DataFrame({'location': 1,
'data': ['3', '4', '5'],
'time': [datetime.datetime(2022,7,16,21,0,0).time(),
datetime.datetime(2022,7,16,21,1,0).time(),
datetime.datetime(2022,7,16,21,3,0).time()]})
df2 = pd.DataFrame({'location': 2,
'data': ['8', '6', '7'],
'time': [datetime.datetime(2022,7,17,21,0,0).time(),
datetime.datetime(2022,7,17,21,2,0).time(),
datetime.datetime(2022,7,17,21,3,0).time()]})
df = pd.concat([df1,df2], axis=0)
fig = px.line(df, x="time", y="data", color='location')
fig.show()
Thanks!
CodePudding user response:
The problem is with the time column. As you convert it to time(), this will be converted to object when you combine the dataframes. Check df.info(). To avoid this, leave the data in datetime format and use update_axis() to let px set the time. Updated code below...
import pandas as pd
import plotly.express as px
df1 = pd.DataFrame({'location': 1,
'data': ['3', '4', '5'],
'time': [datetime.datetime(2022,7,16,21,0,0),
datetime.datetime(2022,7,16,21,1,0),
datetime.datetime(2022,7,16,21,3,0)]})
df2 = pd.DataFrame({'location': 2,
'data': ['8', '6', '7'],
'time': [datetime.datetime(2022,7,16,21,0,0),
datetime.datetime(2022,7,16,21,2,0),
datetime.datetime(2022,7,16,21,3,0)]})
df = pd.concat([df1,df2], axis=0)
fig = px.line(df, x="time", y="data", color='location')
fig.update_xaxes(tickformat="%H:%M:%S")
fig.show()
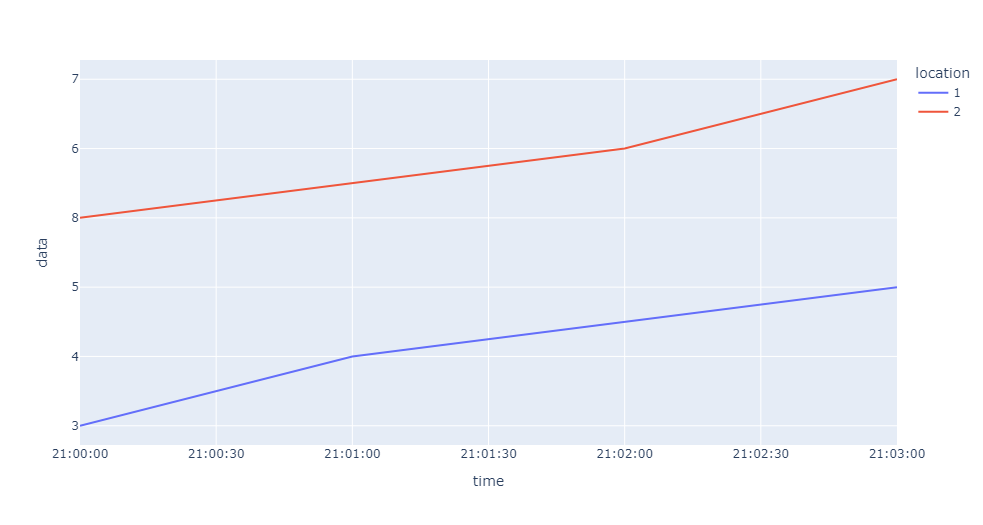
Plot
EDIT
As per your updated requirement, you want to remove the date section and show ONLY time, without caring for the date. To do this, you can go back to taking only the time() in your dataframe creation. Post concatenation of the dfs, use a dummy date (2022-01-01 here) and create the datetime and plot. This will give you below graph.
## Note that you need to use .time()
df1 = pd.DataFrame({'location': 1, 'data': ['3', '4', '5'],
'time': [datetime.datetime(2022,7,17,21,0,0).time(),
datetime.datetime(2022,7,17,21,1,0).time(),
datetime.datetime(2022,7,17,21,3,0).time()]})
df2 = pd.DataFrame({'location': 2, 'data': ['8', '6', '7'],
'time': [datetime.datetime(2022,7,16,21,0,0).time(),
datetime.datetime(2022,7,16,21,2,0).time(),
datetime.datetime(2022,7,16,21,3,0).time()]})
df = pd.concat([df1,df2], axis=0)
date = str(datetime.datetime.strptime('2022-01-01', '%Y-%m-%d').date()) ##Random dummy date
df['time'] = pd.to_datetime(date " " df['time'].astype(str)) ##Convert back to datetime
fig = px.line(df, x="time", y="data", color='location')
fig.update_xaxes(tickformat="%H:%M")
fig.show()
Now, for the second part of your requirement - the graph needs to always show 9PM to 8AM. To do this, you will need to use the range_x with the start and end times. Replace above px.line() with these lines...
dt = datetime.datetime.strptime('2022-01-01', '%Y-%m-%d')
starttime = dt.replace(hour=21, minute=0) ## Start time is 9PM
dt = datetime.datetime.strptime('2022-01-02', '%Y-%m-%d')
endtime = dt.replace(hour=8, minute=0) ## End time is 8AM next day
fig = px.line(df, x="time", y="data", color='location', range_x=[starttime, endtime])
Please note that I have shown both plots to showcase that, if you take a longer period (9 to 8), the lines will just look like vertical lines. Hope you are okay with that.
CodePudding user response:
- started by simulating data that has the features you describe. From 21:00 to 08:00 on different dates and with different randomly removed minutes
- now integrate this data. Have taken approach
- fill missing minutes by outer join to all minutes in each dataframe
- outer join the two data frames on time only
This gives a different struct data frame:
| location_x | time_x | Data_x | t | location_y | time_y | Data_y | |
|---|---|---|---|---|---|---|---|
| 0 | 1 | 2022-09-01 21:00:00 | 0 | 21:00:00 | 2 | 2022-09-04 21:00:00 | 1 |
| 1 | 1 | 2022-09-01 21:01:00 | 0.0302984 | 21:01:00 | 2 | 2022-09-04 21:01:00 | 0.999541 |
| 2 | 1 | 2022-09-01 21:02:00 | 0.060569 | 21:02:00 | 2 | 2022-09-04 21:02:00 | 0.998164 |
| 3 | 1 | 2022-09-01 21:03:00 | 0.0907839 | 21:03:00 | 2 | 2022-09-04 21:03:00 | 0.995871 |
| 4 | 1 | 2022-09-01 21:04:00 | 0.120916 | 21:04:00 | 2 | 2022-09-04 21:04:00 | nan |
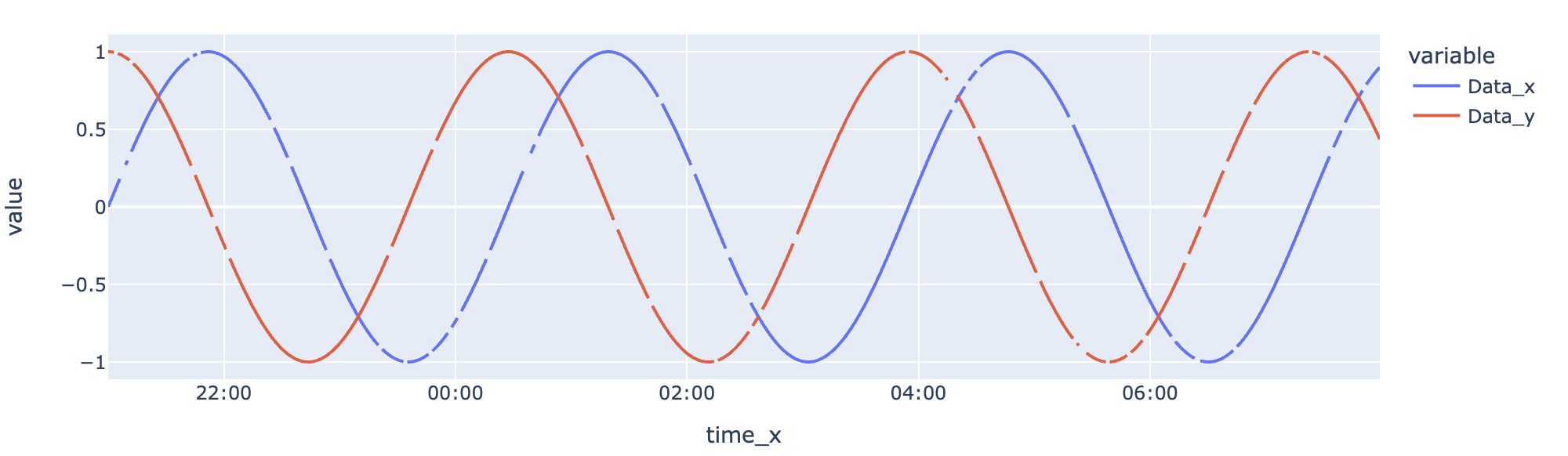
This is then simple to generate a px.line() figure from. Traces being Data_x and Data_y. Have used datetime column time_x for xaxis. This then works well as datetime and continuous axes are well integrated. Updated tickformat so date part of axis is not displayed.
import pandas as pd
import numpy as np
import plotly.express as px
dr = pd.date_range("2022-09-01 21:00", "2022-09-02 08:00", freq="1Min")
# data to match question, two dataframes from 21:00 to 08:00, different dates with some holes
# with different dates
dfs = [
pd.DataFrame(
{
"location": np.full(len(dr), l),
"time": dr pd.DateOffset(days=o),
"Data": f(np.linspace(0, 20, len(dr))),
}
)
.sample(frac=0.95)
.sort_index()
for l, o, f in zip([1, 2], [0, 3], [np.sin, np.cos])
]
df1 = dfs[0]
df2 = dfs[1]
# let's integrate the dataframes
# 1. fill the holes in each dataframe by doing an outer join to all times
# 2. outer join the two dataframes on just the time
df = pd.merge(
*[
pd.merge(
d,
pd.DataFrame(
{"time": pd.date_range(d["time"].min(), d["time"].max(), freq="1min")}
),
on="time",
how="outer",
)
.fillna({"location": l})
.assign(t=lambda d: d["time"].dt.time)
for d, l in zip([df1, df2], [1, 2])
],
on="t",
how="outer",
)
# finally generate plotly line chart using columns created by merging the data
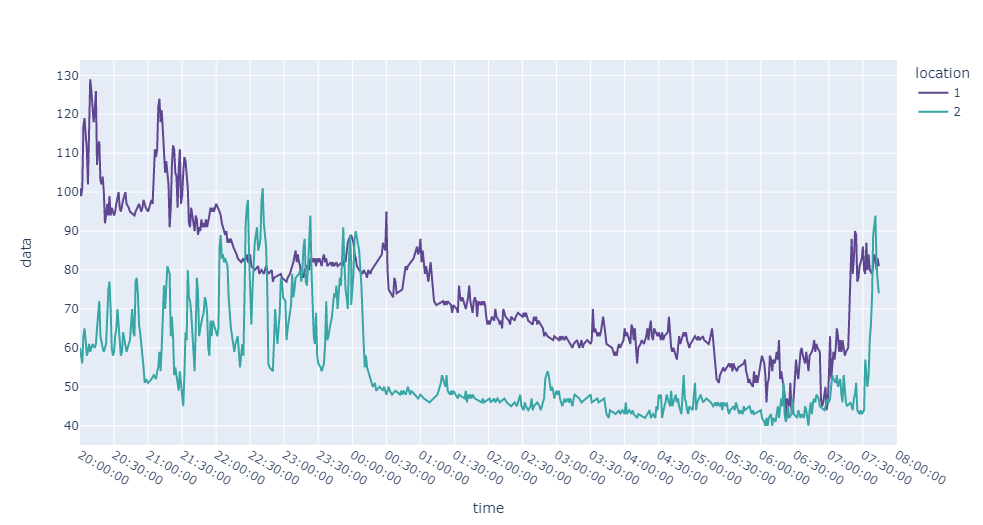
# it's clearly observed there are gaps in both traces
px.line(
df.sort_values("time_x"), x="time_x", y=["Data_x", "Data_y"], hover_data=["time_y"]
).update_layout({"xaxis": {"tickformat": "%H:%M"}})
output
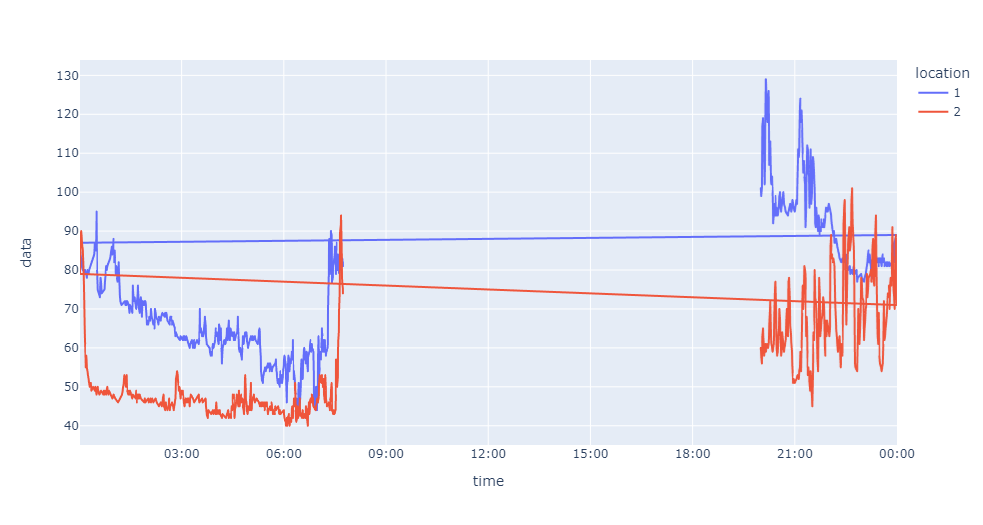
CodePudding user response:
Thank you for your help @Redox it was very helpful but unfortunately doesn't work as I want it to when using the full datasets. This is the result for the equivalent of this:
## Note that you need to use .time()
df1 = pd.DataFrame({'location': 1, 'data': ['3', '4', '5'],
'time': [datetime.datetime(2022,7,17,21,0,0).time(),
datetime.datetime(2022,7,17,21,1,0).time(),
datetime.datetime(2022,7,17,21,3,0).time()]})
df2 = pd.DataFrame({'location': 2, 'data': ['8', '6', '7'],
'time': [datetime.datetime(2022,7,16,21,0,0).time(),
datetime.datetime(2022,7,16,21,2,0).time(),
datetime.datetime(2022,7,16,21,3,0).time()]})
df = pd.concat([df1,df2], axis=0)
date = str(datetime.datetime.strptime('2022-01-01', '%Y-%m-%d').date()) ##Random dummy date
df['time'] = pd.to_datetime(date " " df['time'].astype(str)) ##Convert back to datetime
fig = px.line(df, x="time", y="data", color='location')
fig.update_xaxes(tickformat="%H:%M")
fig.show()
When I try this:
dt = datetime.datetime.strptime('2022-01-01', '%Y-%m-%d')
starttime = dt.replace(hour=21, minute=0) ## Start time is 9PM
dt = datetime.datetime.strptime('2022-01-02', '%Y-%m-%d')
endtime = dt.replace(hour=8, minute=0) ## End time is 8AM next day
fig = px.line(df, x="time", y="data", color='location', range_x=[starttime, endtime])
CodePudding user response:
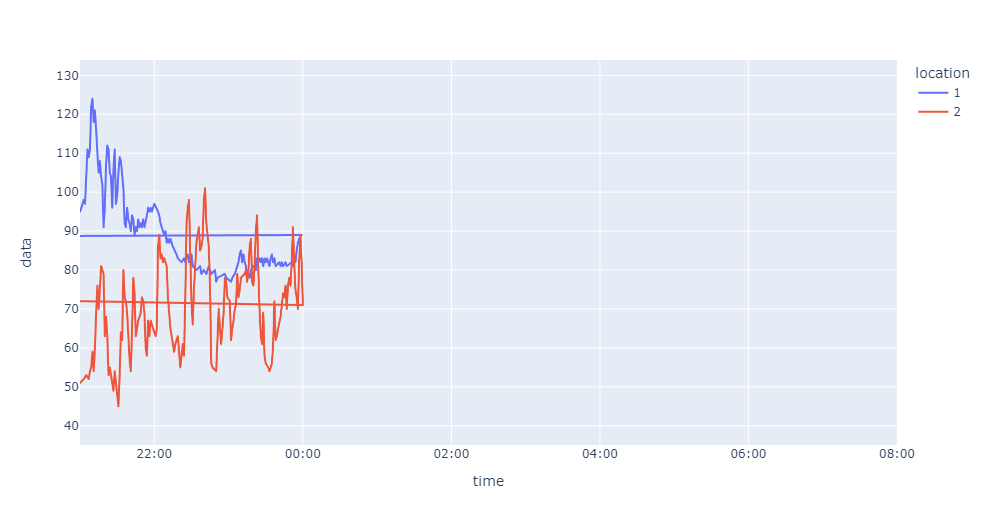
Here is what worked for me eventually:
df1 = pd.DataFrame({'location': 1, 'data': ['3', '4', '5'],
'time_num': [datetime.datetime(2022,7,17,21,0,0).time().hour datetime.datetime(2022,7,17,21,0,0).time().minute/60,
datetime.datetime(2022,7,17,21,1,0).time().hour datetime.datetime(2022,7,17,21,0,0).time().minute/60,
datetime.datetime(2022,7,17,21,3,0).time().hour datetime.datetime(2022,7,17,21,0,0).time().minute/60, ]})
df2 = pd.DataFrame({'location': 2, 'data': ['8', '6', '7'],
'time_num': [datetime.datetime(2022,7,16,21,0,0).time().hour datetime.datetime(2022,7,16,21,0,0).time().minute/60,
datetime.datetime(2022,7,16,21,2,0).time().hour datetime.datetime(2022,7,16,21,2,0).time().minute/60,
datetime.datetime(2022,7,16,21,3,0).time().hour datetime.datetime(2022,7,16,21,3,0).time().minute/60]})
df_skeleton = pd.DataFrame()
df_skeleton['date'] = pd.date_range(datetime.datetime(2022,7,16,20,0,0), datetime.datetime(2022,7,17,8,0,0), freq = '1min')
df_skeleton['time']=df_test['date'].dt.strftime('%H:%M:%S')
df_skeleton['hour']=df_test['date'].dt.strftime('%H')
df_skeleton['min']=df_test['date'].dt.strftime('%M')
df_skeleton[['hour', 'min']] = df_test[['hour', 'min']].astype(int)
df_skeleton['time_num'] = df_test['hour'] df_test['min']/60
result_1 = pd.merge(df_skeleton, df1, how="left", on=["time_num", "time_num"])
result_2 = pd.merge(df_skeleton, df2, how="left", on=["time_num", "time_num"])
result_1['location'] = '1'
fig = px.line(result_1, x='time', y='data',color='location')
fig.add_scatter(x=result_2['time'], y=result_2['data'],mode='lines', name='2')
fig.update_traces(connectgaps=True)
fig.show()
I'm not overly pleased with it but it works both with the dummy dataframes and the full dataframes.