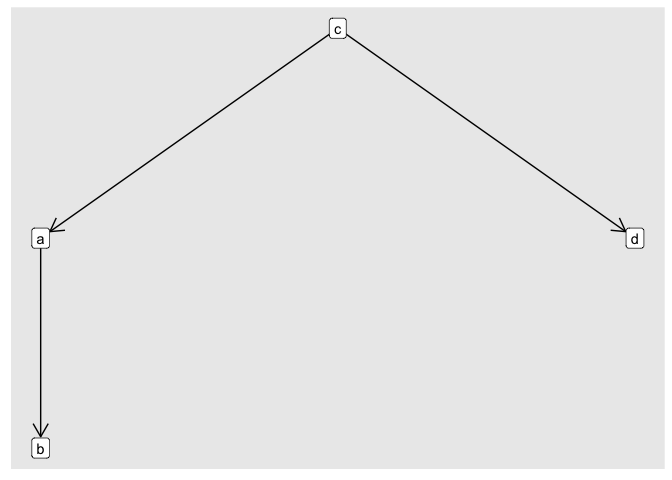
I want to create a directed network visualization with the X-Axis fixed in time.
Assuming I have the following data:
Nodes
| ID | Label | Date |
|---|---|---|
| 1 | a | 2022-02-01 |
| 2 | b | 2022-03-13 |
| 3 | c | 2022-03-22 |
| 4 | d | 2022-04-20 |
Edges
| From | To |
|---|---|
| 1 | 2 |
| 3 | 1 |
| 3 | 4 |
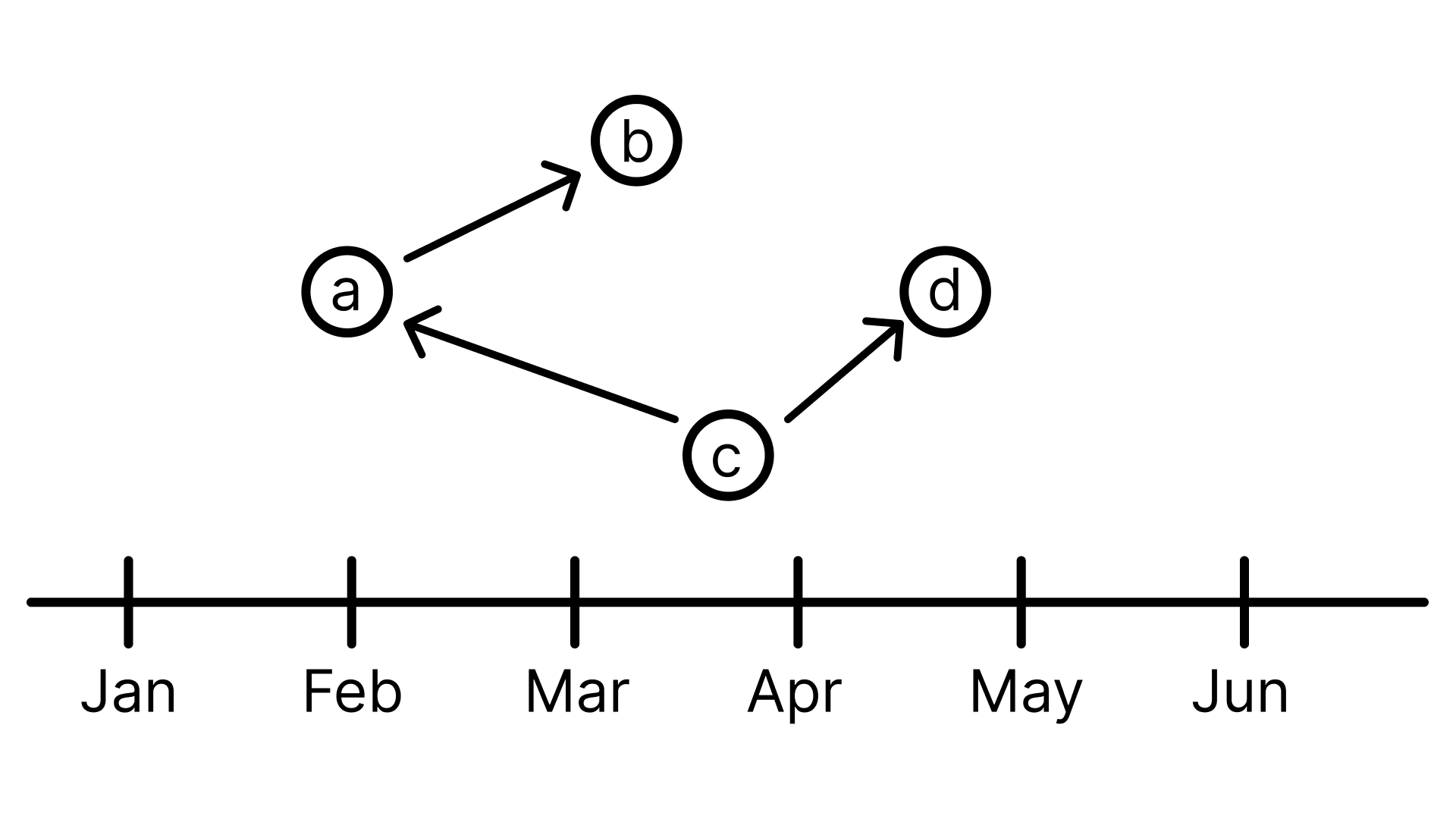
I want to create a visualization which looks like this:
I tried using ggraph but I am having difficulty fixing the X-Axis based on the date.
Any advice is welcome!
Below is some starter code:
library(tidyverse)
library(tidygraph)
#>
#> Attaching package: 'tidygraph'
#> The following object is masked from 'package:stats':
#>
#> filter
library(ggraph)
nodes <- tibble(id=1:4,label=letters[1:4], date=as.Date(c("2022-02-01",
"2022-03-13",
"2022-03-22",
"2022-04-20")))
nodes
#> # A tibble: 4 × 3
#> id label date
#> <int> <chr> <date>
#> 1 1 a 2022-02-01
#> 2 2 b 2022-03-13
#> 3 3 c 2022-03-22
#> 4 4 d 2022-04-20
edges <- tibble(from = c(1L, 3L, 3L),
to = c(2L, 1L, 4L))
edges
#> # A tibble: 3 × 2
#> from to
#> <int> <int>
#> 1 1 2
#> 2 3 1
#> 3 3 4
graph <- tbl_graph(nodes = nodes, edges = edges, directed=TRUE)
graph
#> # A tbl_graph: 4 nodes and 3 edges
#> #
#> # A rooted tree
#> #
#> # Node Data: 4 × 3 (active)
#> id label date
#> <int> <chr> <date>
#> 1 1 a 2022-02-01
#> 2 2 b 2022-03-13
#> 3 3 c 2022-03-22
#> 4 4 d 2022-04-20
#> #
#> # Edge Data: 3 × 2
#> from to
#> <int> <int>
#> 1 1 2
#> 2 3 1
#> 3 3 4
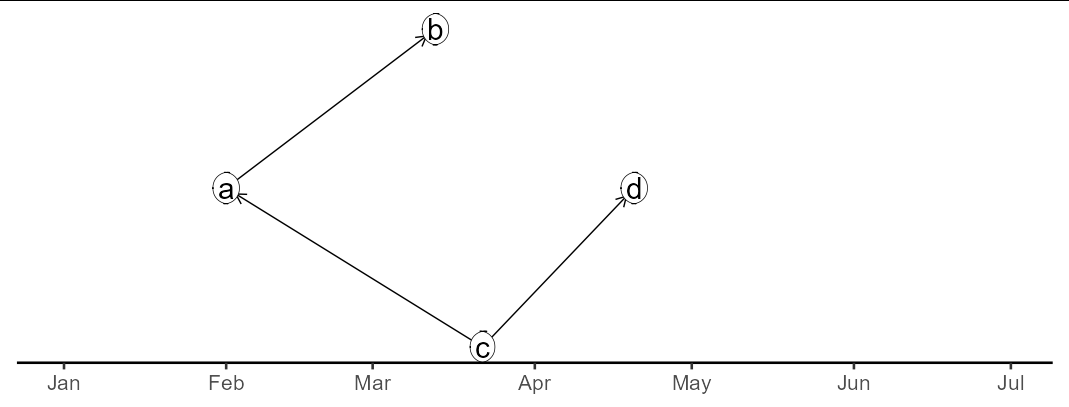
ggraph(graph)
geom_edge_link(end_cap = circle(3, 'mm'),
arrow = arrow(length = unit(4, 'mm')))
geom_node_label(aes(label = label))
#> Using `tree` as default layout