I created a geom_histogram using the dataset and code below, and I wanted to label each bar in histogram with the subject ID and color the bar according to the metabolizer group, I noticed that for some reason the ID label and the color don't match, the ID is correct on the x-axis value but it is not colored according to the group.
For example ID 72 in the graph below has a value of -2.85, the ID is correct on the x-axis location but should be colored dark green as a PM, same for ID 33 should be UM light blue color and so on!
Any suggestions! Thanks
The dataset:
Set.seed(4)
df <- data.frame(ID = factor(1:72), gengroup = c("UM","NM" ,"IM", "PM"), value = 2 - rgamma(72, 3, 2))
Histogram code:
p1 <- ggplot(df, aes(x = value, fill = gengroup))
scale_fill_brewer(aes(name= "Metabolizer group"), palette = "Paired", labels= c("UM","NM" ,"IM", "PM"))
geom_histogram(bins = 30)
stat_bin(geom = "text", bins = 30,size =2, na.rm = TRUE,
aes(label = ifelse(after_stat(count) == 0, NA, after_stat(group)),
group = ID, y = after_stat(count)),
position = position_stack(vjust = 0.5))
labs(x = NULL)
show(p1)
Graph:
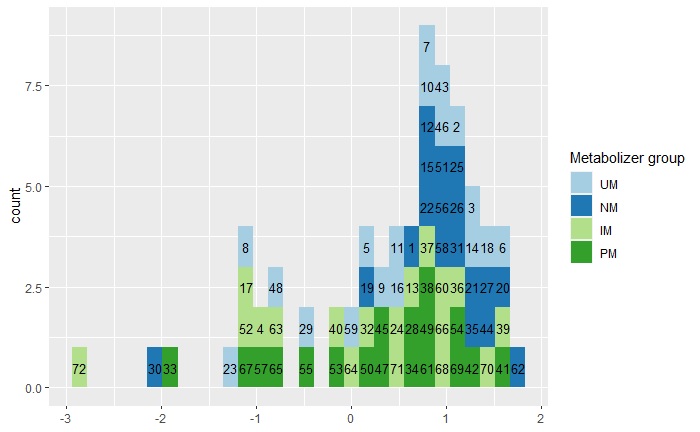
CodePudding user response:
You could extract the colors of the Paired palette using brewer.pal from RColorBrewer and manually assign them with scale_fill_manual like this:
set.seed(4)
df <- data.frame(ID = factor(1:72), gengroup = c("UM","NM" ,"IM", "PM"), value = 2 - rgamma(72, 3, 2))
library(ggplot2)
library(RColorBrewer)
colors <- brewer.pal(4, "Paired")
p1 <- ggplot(df, aes(x = value, fill = gengroup))
geom_histogram(bins = 30)
stat_bin(geom = "text", bins = 30,size =2, na.rm = TRUE,
aes(label = ifelse(after_stat(count) == 0, NA, after_stat(group)),
group = ID, y = after_stat(count)),
position = position_stack(vjust = 0.5))
scale_fill_manual("Metabolizer group", values = c("UM" = colors[1],
"NM" = colors[2],
"IM" = colors[3],
"PM" = colors[4]))
labs(x = NULL)
show(p1)
Created on 2022-09-12 with reprex v2.0.2
CodePudding user response:
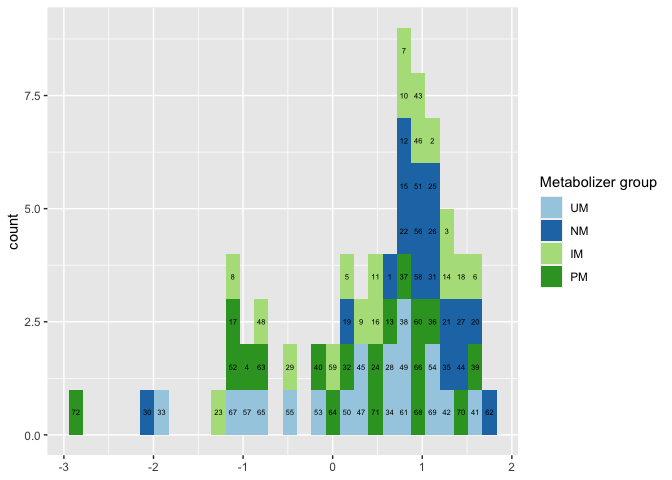
There are actually two issues in your code:
- Using
labels= c("UM","NM" ,"IM", "PM")you are changing the labels for your groups in the legend. But under the hood the colors are assigned by the order of the groups in the data, which by default isc("IM","NM" ,"PM", "UM"), e.g. the dark green which is labelledPMis actually assigned to gengroupUM. To fix that you set thelimits = c("UM", "NM", "IM", "PM")instead of usinglabels
set.seed(4)
df <- data.frame(ID = factor(1:72), gengroup = c("UM", "NM", "IM", "PM"), value = 2 - rgamma(72, 3, 2))
library(dplyr)
library(ggplot2)
ggplot(df, aes(x = value, fill = gengroup))
scale_fill_brewer(aes(name= "Metabolizer group"), palette = "Paired", limits = c("UM", "NM", "IM", "PM"))
geom_histogram(bins = 30)
stat_bin(geom = "text", bins = 30,size =2, na.rm = TRUE,
aes(label = ifelse(after_stat(count) == 0, NA, after_stat(group)),
group = ID, y = after_stat(count)),
position = position_stack(vjust = 0.5))
labs(x = NULL)
As you see now your ID 72 get the correct dark green and the ID 33 the light blue.
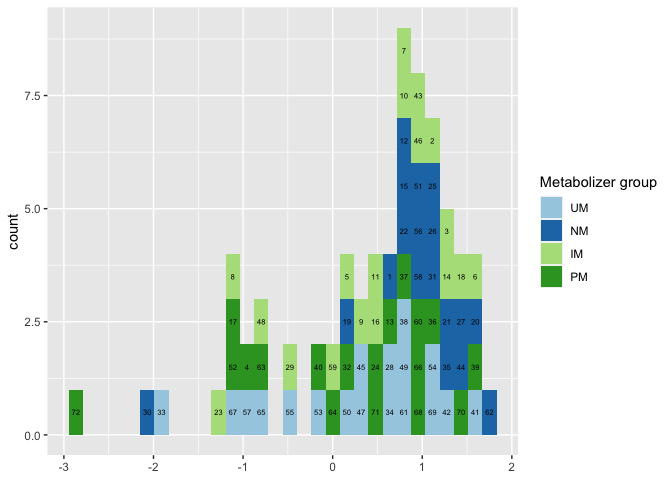
However there are still some issues in all cases where the bars contain more than one ID, e.g. ID 8 should also be colored dark green but is colored light green.
The reason for that is that you apply a different grouping for the geom_histogram and for adding the labels via stat_bin. For the first the grouping is defined by gengroup while for the second you group by ID. This could be seen clearly by grouping the geom_histogram by ID too:
ggplot(df, aes(x = value, fill = gengroup))
scale_fill_brewer(aes(name= "Metabolizer group"), palette = "Paired", limits = c("UM", "NM", "IM", "PM"))
geom_histogram(aes(group = ID), bins = 30)
stat_bin(geom = "text", bins = 30,size =2, na.rm = TRUE,
aes(label = ifelse(after_stat(count) == 0, NA, after_stat(group)),
group = ID, y = after_stat(count)),
position = position_stack(vjust = 0.5))
labs(x = NULL)
As can be seen now we get the right colors but the bars are no longer stacked in the order of gengroup
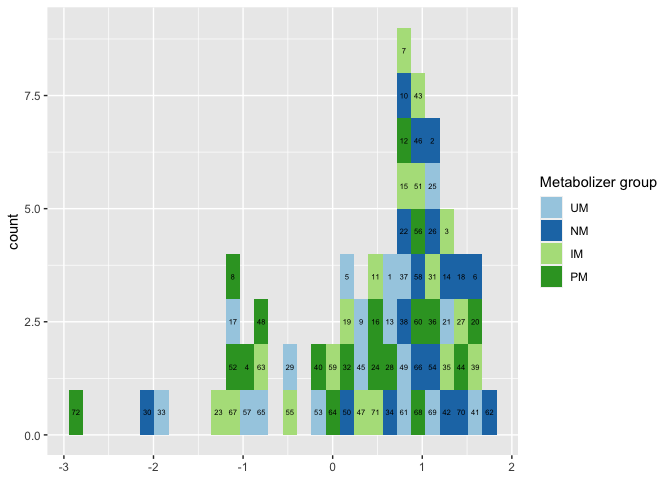
- To fix that and to stack the labels by
gengroupyou could convertIDto afactorwith the order of the IDs set according to the order ofgengroup. To this end I arrange the data first and useforcats::fct_inorder. However, to get right labels we also have to make use of a lookup table to assign the right labels insideafter_stat:
df <- df |>
arrange(gengroup) |>
mutate(ID = forcats::fct_inorder(ID))
labels <- setNames(levels(df$ID), seq_along(levels(df$ID)))
ggplot(df, aes(x = value, fill = gengroup))
scale_fill_brewer(aes(name = "Metabolizer group"), palette = "Paired", limits = c("UM", "NM", "IM", "PM"))
geom_histogram(bins = 30)
stat_bin(
geom = "text", bins = 30, size = 2, na.rm = TRUE,
aes(
label = ifelse(after_stat(count) == 0, NA, after_stat(labels[group])),
group = ID, y = after_stat(count)
),
position = position_stack(vjust = 0.5)
)
labs(x = NULL)

