I am trying to simulate high dimension multivariate normal data in R with n = 100, and p = 400 (two different groups of variables with some correlations). Below are my codes:
## load library MASS
library(MASS)
## sample size set to n = 100
sample_size <- 100
## I try to simulate two different groups of variables for each with 200 variables
sample_meanvector <- c(runif(200,0,1), runif(200,6,8))
## covariance matrix, some variables set to be correlated
sample_covariance_matrix <- matrix(NA, nrow = 400, ncol = 400)
diag(sample_covariance_matrix) <- 1
set.seed(666)
sample_covariance_matrix[lower.tri(sample_covariance_matrix)] <- runif(79800, 0.00001, 0.2)
sample_covariance_matrix[lower.tri(sample_covariance_matrix)][sample(1:79800, 10000)] <- runif(10000, 0.6, 0.9)
## make the matrix symmetric
sample_covariance_matrix[upper.tri(sample_covariance_matrix)]<-t(sample_covariance_matrix)[upper.tri(sample_covariance_matrix)]
## create multivariate normal distribution
sample_distribution <- mvrnorm(n = sample_size,
mu = sample_meanvector,
Sigma = sample_covariance_matrix)
However, every time I run this mvrnorm function, I got the error:
Error in mvrnorm(n = sample_size, mu = sample_meanvector, Sigma = sample_covariance_matrix) : 'Sigma' is not positive definite
I have two questions:
- Why do I have this error?
- How can I edit my codes to simulate the high dimension multivariate normal data following my idea mentioned above?
Thanks much!
CodePudding user response:
Here is an approach that can generate a highly correlated matrix :
library(MASS)
library(Matrix)
sample_size <- 100
sample_meanvector <- c(runif(200,0,1), runif(200,6,8))
sample_covariance_matrix <- matrix(NA, nrow = 400, ncol = 400)
diag(sample_covariance_matrix) <- 1
set.seed(666)
mat_Sim <- matrix(data = NA, nrow = 400, ncol = 400)
U <- runif(n = 400) * 0.5
for(i in 1 : 400)
{
if(i <= 350)
{
U_Star <- pmin(U 0.25 * runif(n = 400), 0.99999)
}else
{
U_Star <- pmin(pmax(U sample(c(-1, 1), size = 400, replace = TRUE) * runif(n = 400), 0.00001), 0.99999)
}
mat_Sim[, i] <- qnorm(U_Star)
}
cor_Mat <- cor(mat_Sim)
sample_covariance_matrix <- cor_Mat * 200
## create multivariate normal distribution
sample_distribution <- mvrnorm(n = sample_size,
mu = sample_meanvector,
Sigma = sample_covariance_matrix)
image(cor_Mat)
with 7 / 8 of the matrix correlated between 0.6 and 0.8 and 1 / 8 of the matrix correlated between 0 and 0.3. 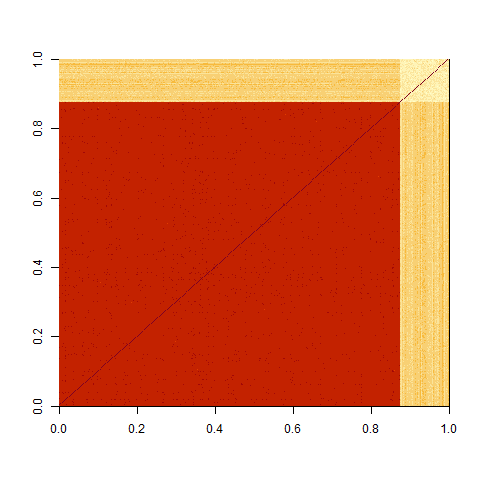
CodePudding user response:
You can consider the following code :
library(MASS)
library(Matrix)
sample_size <- 100
sample_meanvector <- c(runif(200,0,1), runif(200,6,8))
sample_covariance_matrix <- matrix(NA, nrow = 400, ncol = 400)
diag(sample_covariance_matrix) <- 1
set.seed(666)
sample_covariance_matrix[lower.tri(sample_covariance_matrix)] <- runif(79800, 0.00001, 0.2)
sample_covariance_matrix[lower.tri(sample_covariance_matrix)][sample(1:79800, 10000)] <- runif(10000, 0.6, 0.9)
sample_covariance_matrix[upper.tri(sample_covariance_matrix)]<-t(sample_covariance_matrix)[upper.tri(sample_covariance_matrix)]
sample_covariance_matrix_Near_PD <- nearPD(sample_covariance_matrix)$mat
## create multivariate normal distribution
sample_distribution <- mvrnorm(n = sample_size,
mu = sample_meanvector,
Sigma = sample_covariance_matrix_Near_PD)
